About FusionEncoder
Intrinsic disorder regions (IDRs) play a significant role in various biological processes and are widely distributed in proteins. Thus, accurately predicting these regions is essential for analyzing protein structure and function. Amino acid feature extraction is a pivotal step in the development of computational predictive models. Existing methods typically rely on traditional biological features (e.g., PSSM) or use pre-trained protein language models (PPLMs) to extract sequence semantic information, often resorting to straightforward feature concatenation. However, these approaches fail to capture the multi-semantic interactions between traditional biological features and PPLMs-based features. In this study, we present a method named FusionEncoder designed for the integration of traditional biological and PPLMs-based features of the protein. FusionEncoder is a fusion network built on a variant of long short-term memory(LSTM). We consider traditional biological features and PPLMs-based features to be two types of semantic inputs within a “multi-semantic” space. Traditional features are input into the cell state of the LSTM, while PPLMs-based features are fed into the input part. A fusion cell is then utilized to fuse these two types of features. This strategy leverages the capability of LSTM to encode long sequences, enhancing context-aware semantic learning of amino acid sequences. Finally, a transformer-based encoder layer is employed to predict the IDRs. Experimental results on four independent test datasets showed that FusionEncoder significantly improves the accuracy of amino acid feature representation and outperforms other compared methods. .
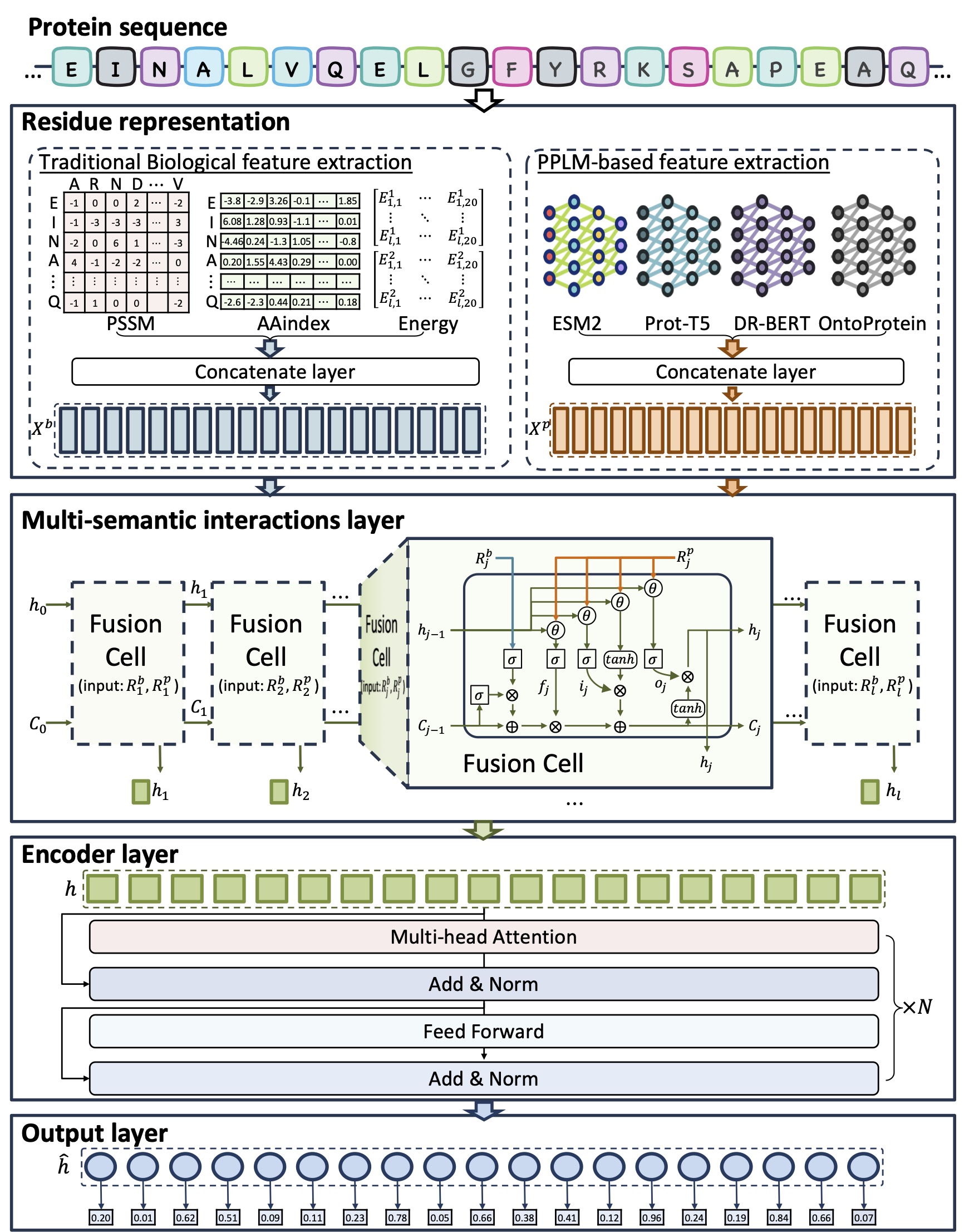
The framework of FusionEncoder. (a) The residue feature extraction process. This stage involves traditional biological feature extraction (PSSM, AAindex, and energy-based methods) and PPLMs-based feature extraction(ESM, ProtT5, DR-BERT, and OntoProtein methods). (b) Multi-Semantic Interaction Layer. In this layer, traditional biological features and PPLMs-based features are fused using the FusionCell module, while long short-term memory (LSTM) facilitates information transfer between residues. (c) Encoder Layer. A Transformer-based encoder module is employed to encode protein sequences. (d) The output layer is utilized to perform predictions for IDRs.