Intrinsically disordered proteins (IDPs) and regions (IDRs) are a class of functionally important proteins and regions that lack stable three-dimensional structures under the native physiologic conditions. They participate in critical biological processes and thus are associated with pathogenesis of many severe human diseases. Identifying the IDPs/IDRs and their functions will be helpful for a comprehensive understanding of protein structures and functions, and inform studies of rational drug design. Over the past decades, the exponential growth in the number of proteins with sequence information has deepened the gap between uncharacterized and annotated disordered sequences. Protein language models have recently demonstrated their powerful abilities to capture complex structural and functional information from the enormous quantity of unlabelled protein sequences, providing the opportunities to apply protein language models to uncover the intrinsic disorders and their biological properties from the amino acid sequences. In this study, we proposed a computational predictor called IDP-LM for predicting intrinsic disorder and disorder functions by leveraging the pre-trained protein language models. IDP-LM takes the embeddings extracted from three pre-trained protein language models as the exclusive inputs, including ProtBERT, ProtT5 and a disorder specific language model (IDP-BERT). The ablation analysis shown that the IDP-BERT provided fine-grained feature representations of disorder, and the combination of three language models is the key to the performance improvement of IDP-LM. The evaluation results on independent test datasets demonstrated that the IDP-LM provided high-quality prediction results for both intrinsic disorder and four common disordered functions.

Figure.1 Overview of IDP-LM predictor. The input sequences were processed by three language models to generate the embedding vector for each residue of proteins. The IDP-BERT disordered language model adopts the BERT architecture of stacking multiple Transformer encoders, and it was self-supervised pre-trained with the sequence data collected from the MobiDB and PDB database. Three prediction layers in IDP-LM were used to calculate per-residue disordered propensity scores based on embeddings extracted from three language models, respectively. Then the model outputs the final propensity scores and binary results by fusing the calculations from three prediction layers.
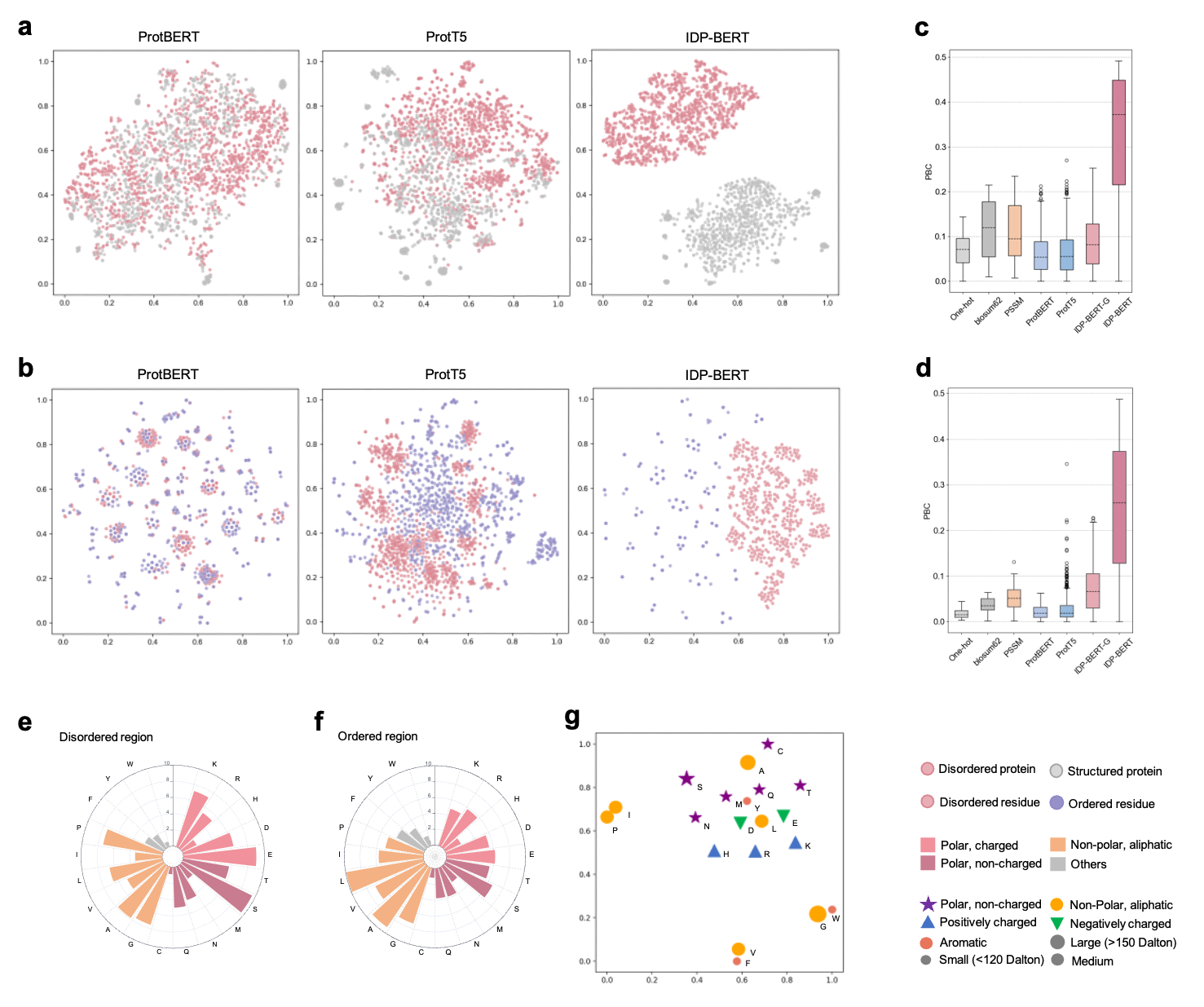
Figure.2 Protein language models capture disordered properties. t-SNE projection visualization of disordered/structured proteins’ (a) and residues’ (b) embedding vectors extracted by three pre-trained protein language models, where the language model trained for disordered proteins (IDP-BERT) learned more fine-grained distinctions of disorder and order. The comparation of point-biserial correlation (PBC) scores calculated based on different feature representations of disordered proteins (c) and residues (d). We included template-free features (One-hot and blosum62), multiple sequence alignment based feature (PSSM), and pre-trained language model encodings (ProtBERT, ProtT5, IDP-BERT-G and IDP-BERT), where the IDP-BERT-G represents the features extracted from the IDP-BERT pretrained with the general mask language modelling. Higher PBC value reflects the information provided by features more relevant with disorder. According to our statistics in the DisProt database, disordered regions (e) are rich in polar residues compared with the ordered regions (f). (g), the t-SNE projections of amino acids encoding vectors captured by IDP-BERT in 2D space conform with their biochemical properties.
We acknowledge with thanks the following databases and softwares used in this server:
DisProt: database of intrinsically disordered proteins.
MobiDB: database of protein disorder and mobility annotations.
PDB: RCSB Protein Data Bank.
ProtTrans: Protein pre-trained language models.
Upon the usage the users are requested to use the following citation:
· Yihe Pang, Bin Liu. IDP-LM: prediction of protein intrinsic disorder and disorder functions based on language models (Submitted)