Introduction
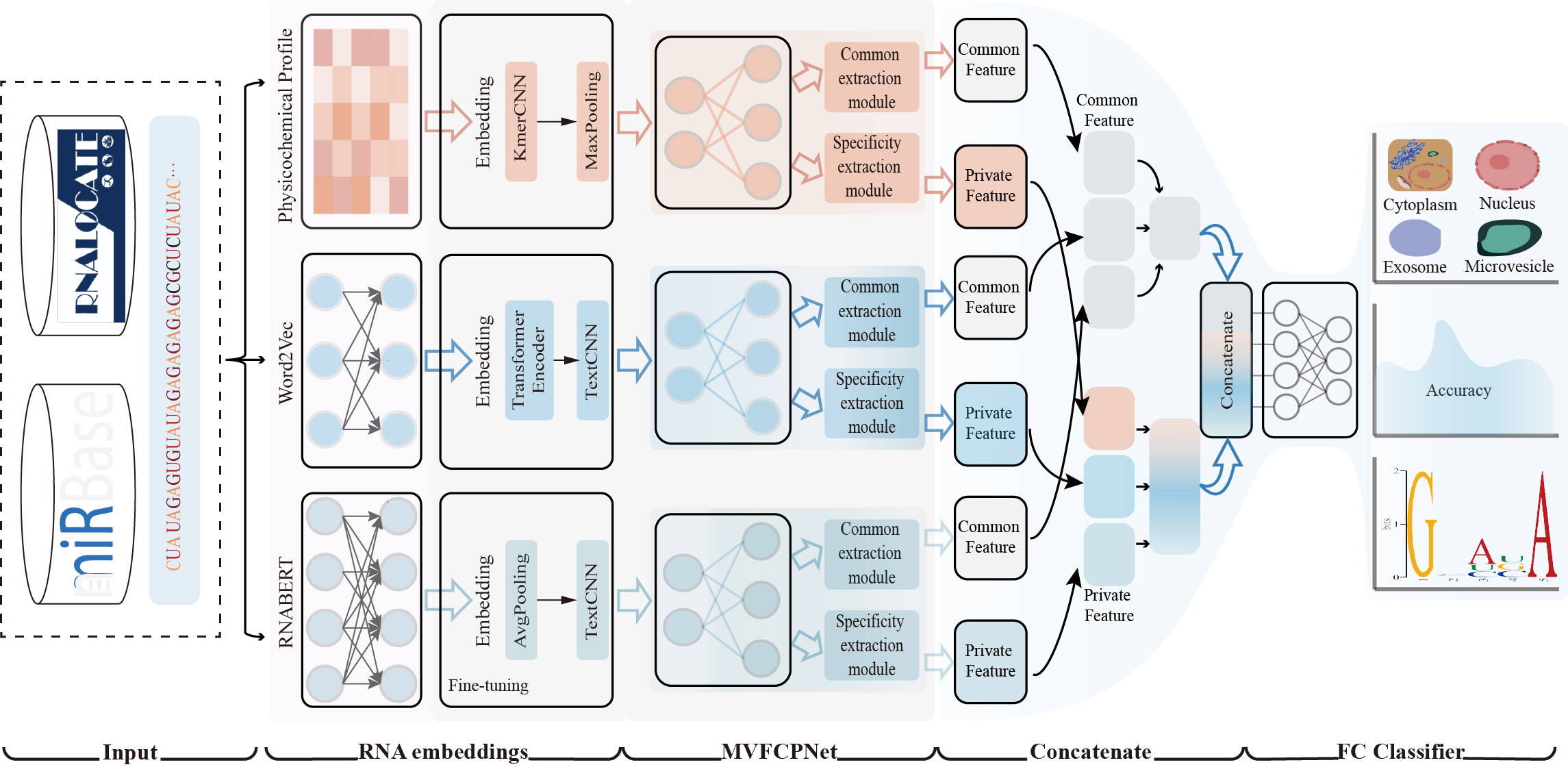
Inspired by multi-view multi-label learning strategy, we proposed a computational method named MMLmiRLocNet for predicting the subcellular localizations of miRNAs. The MMLmiRLocNet predictor extracts diverse feature representations of biological sequences through an investigation into lexical, syntactic, and semantic aspects. Specifically, it integrates lexical attributes derived from k-mer physicochemical profiles, syntactic characteristics obtained via word2vec embeddings, and semantic representations generated by RNABERT feature embeddings. Finally, the multi-view consensus features and specific features extract network constructed to extract consensus features and specific features from various perspectives, respectively. The full connection networks are utilized as the output module to predict the miRNA subcellular localization. The results show that the MMLmiRLocNet outperforms previous methods in terms of F1 score, subACC, and Accuracy, and achieves highly comparable performance with the help of multi-view consensus features and specific features extract network.