About PHR-search
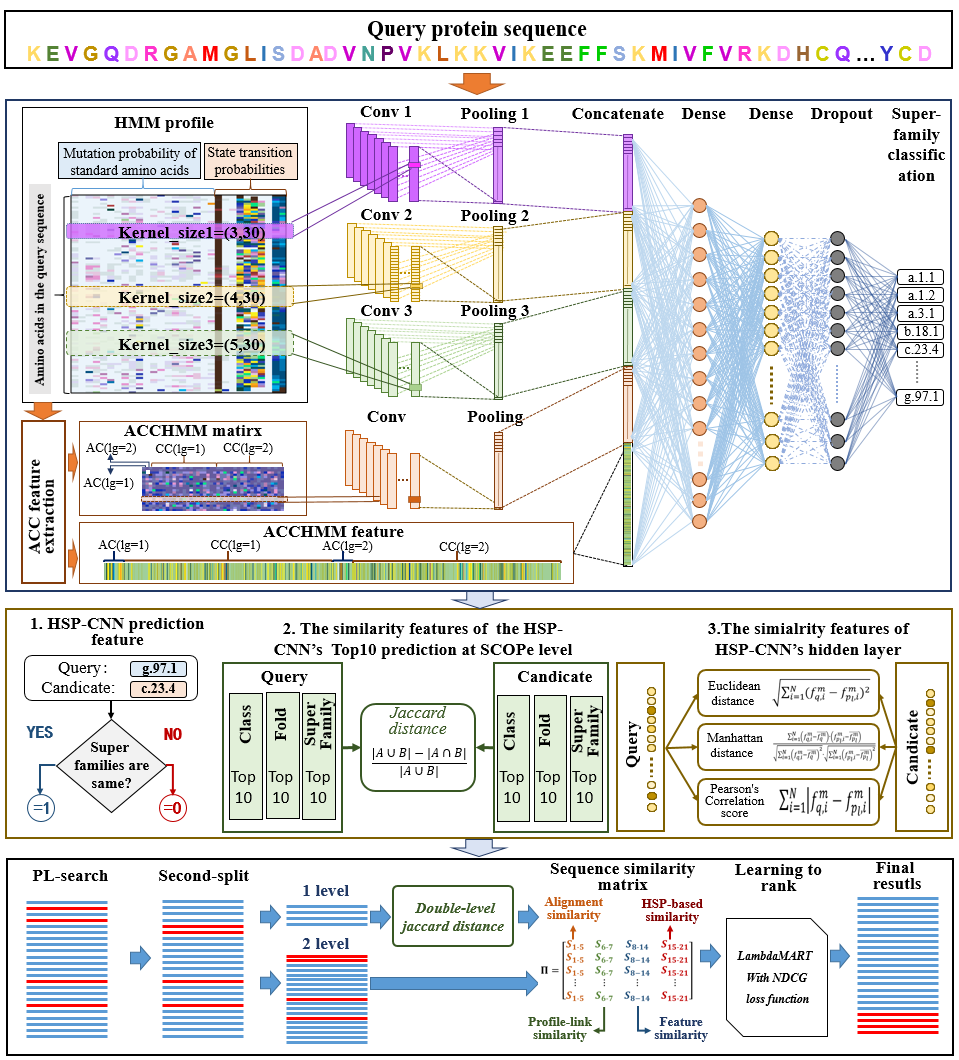
Figure 1. The flowchart of PHR-search.
PHR-search: A novel supervised search framework based on PSI-BLAST for protein remote homology detection
- Motivation: Protein remote homology detection is one of the most fundamental research for protein structure and function prediction. Most search methods for protein remote homology detection are evaluated on the Structural Classification of Proteins-extended (SCOPe) benchmark, but the diversity hierarchical structure relationships between the query protein and candidate proteins are ignored by these methods.
- Results: In order to further improve the predictive performance for protein remote homology detection, a search framework based on the predicted protein hierarchical relationships (PHR-search) is proposed. In the HSP-search framework, the superfamily level prediction information is obtained by extracting the local and global features of Hidden Markov Model (HMM) profile through a convolution neural network, and it is converted to fold level and class level prediction information according to the predicted hierarchical relationships of SCOPe. Based on these predicted protein hierarchical relationships, filtering strategy and re-ranking strategy are used to construct the two-level search of HSP-search. Experimental results show that the PHR-search framework achieves the state-of-the-art performance by employing five basic search methods, including HHblits, JackHMMER, PSI-BLAST, DELTA-BLAST and PSI-BLASTexB.
The non-homology proteins in different levels of SCOPe
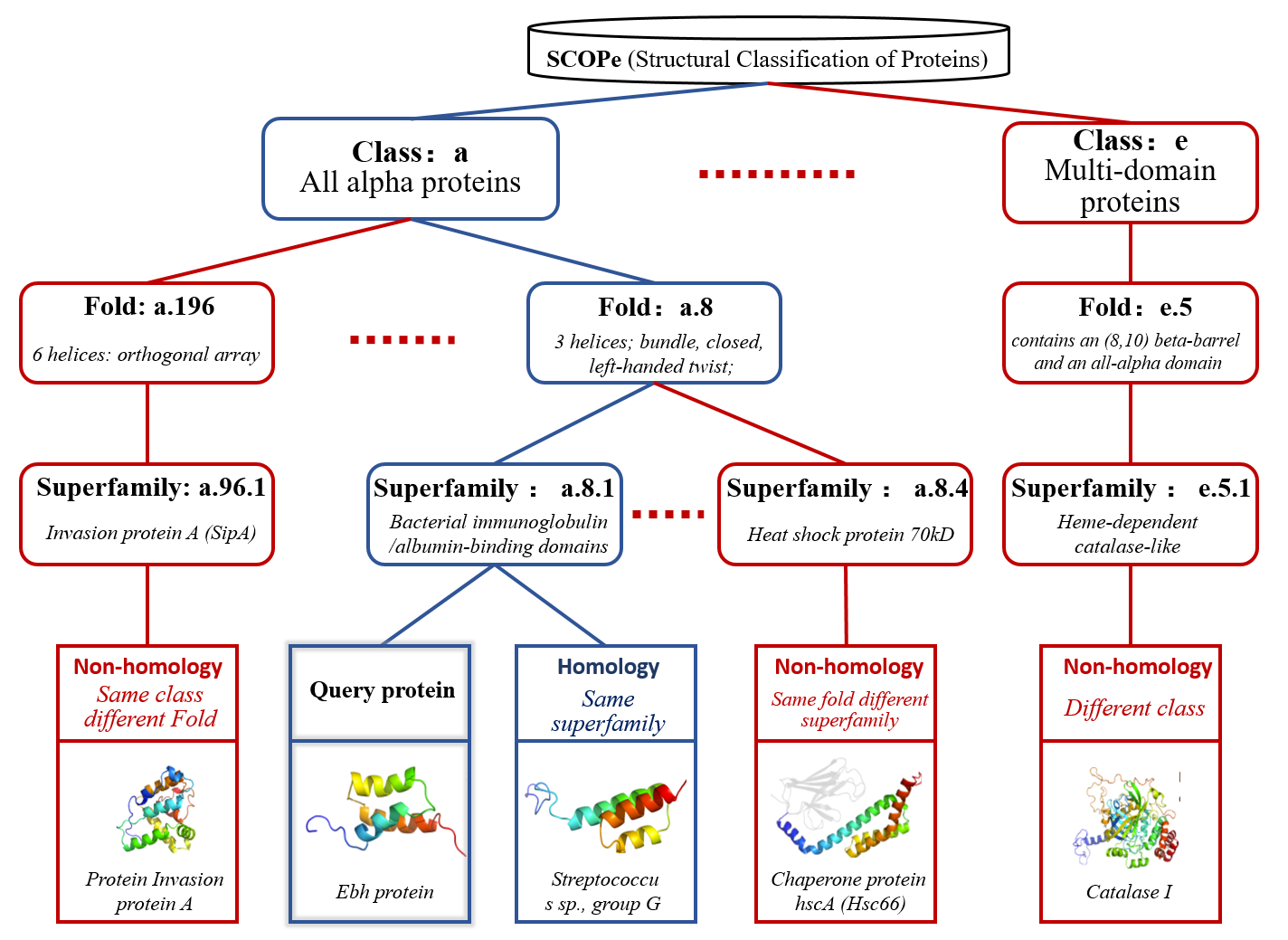
Figure 2. The non-homology proteins in different levels of SCOPe.