 |
PreHom-PCLM:PreHom-PCLM: Protein Remote Homology Detection by Combing Motifs and Protein Cubic Language Model |
 |
PreHom-PCLM:PreHom-PCLM: Protein Remote Homology Detection by Combing Motifs and Protein Cubic Language Model |
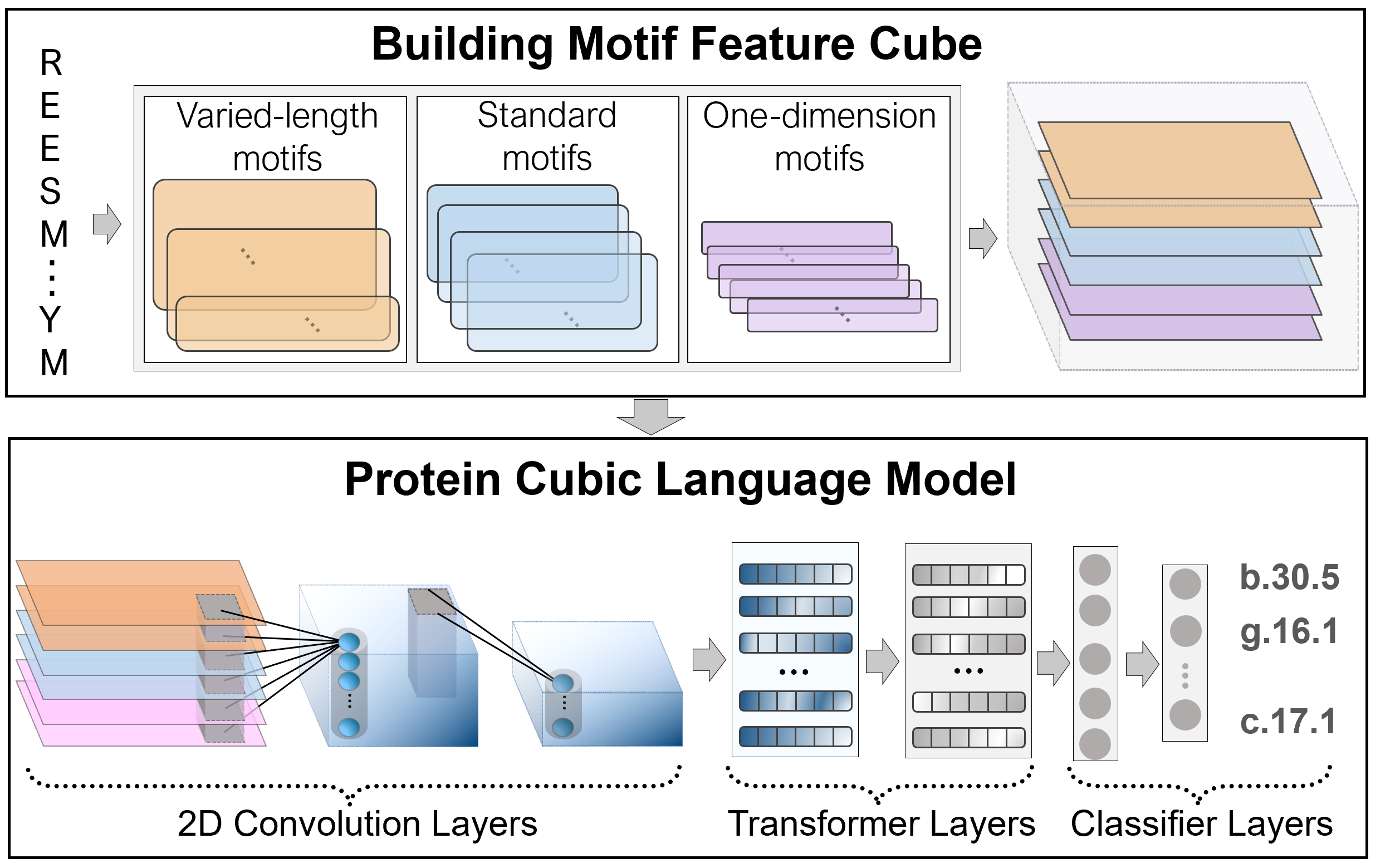
Motivation : Protein remote homology detection is essential for structure prediction, function prediction, disease mechanism understanding, etc. The remote homology relationship depends on multiple protein properties, such as structural information and local sequence patterns. Previous studies have shown the challenges for predicting remote homology relationship by protein features at sequence level (e.g., position specific score matrix). Protein motifs have been used in structure and function analysis due to their unique sequence patterns and implied structural information. Therefore, designing a usable architecture to fuse multiple protein properties based on motifs is urgently needed to improve protein remote homology detection performance. Results: To make full use of the characteristics of motifs, we employed the language model called the Protein Cubic Language Model (PCLM). It combines multiple properties by constructing a motif-based neural network. Based on the PCLM, we proposed a predictor called PreHom-PCLM by extracting and fusing multiple motif features for protein remote homology detection. PreHom-PCLM outperforms the other state-of-the-art methods on the test set and independent test set. Experimental results further prove the effectiveness of multiple features fused by PreHom-PCLM for remote homology detection. Furthermore, the protein features derived from the PreHom-PCLM show strong discriminative power for proteins from different structural classes in the high-dimensional space.
Results : We propose a novel deep neural network-based language model, the Protein Cubic Language Model (PCLM), with three styles of motifs combing. The protein cubic language model integrates different protein properties to identify the remote homology relationship, just like restoring the original scene from multiple photos. The evaluation result on the test set and independent test set shows an outperformed prediction of PCLM than other state-of-the-art methods. Furthermore, sequence representation generated by the PCLM distinguishes proteins into different structural classes at the high-dimensional space.
CONTACT US
Bin Liu
bliu@bliulab.net
School of Computer Science and Technology, Beijing Institute of Technology, China.
Copyright@ By Liu Lab, Beijing Institute of Technology.
网站备案号: 粤ICP备19041859号-1