About TransDFL
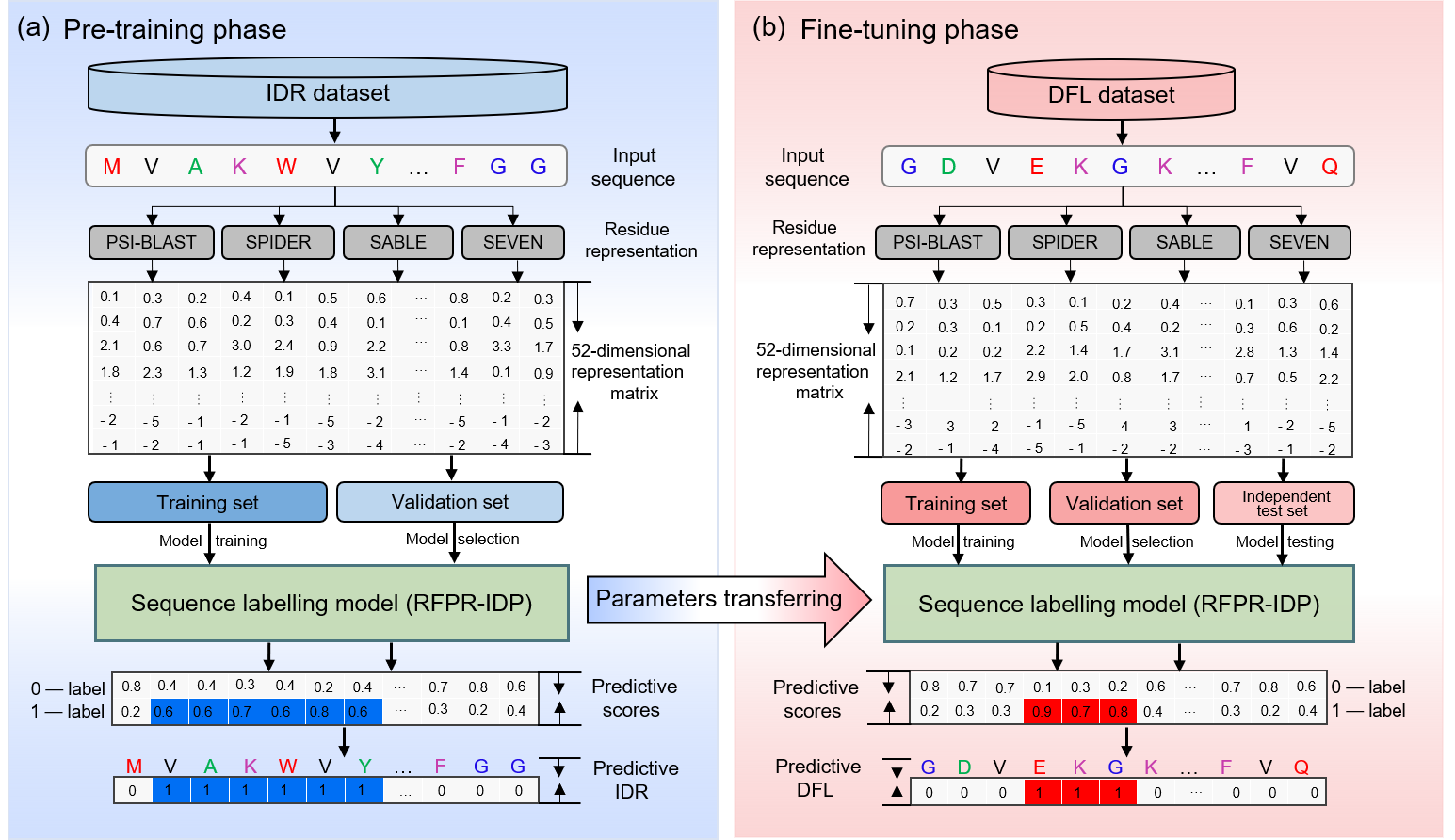
Disordered flexible linkers (DFLs) are the disordered regions of proteins with high level of flexibility, which join different functional domains into a single unit. Computational identification of DFLs is crucial for understanding the functions of intrinsically disordered regions (IDRs). Although several computational predictors have been proposed for identifying DFLs only based on the sequence information. However, they were trained and evaluated with the annotated disordered regions, ignoring the fact that the information of disordered regions is not always available. As a result, DFL predictors tend to predict the ordered residues as DFLs, leading to high false-positive rate (FPR) and low prediction accuracy. DFLs are the extremely flexible regions which are usually predicted as high confident disordered residues [P(D) > 0.9] by an IDR predictor, providing an opportunity to more accurately predict the DFLs via transferring an IDR predictor to a DFL predictor.
In this study, we proposed a new predictor called TransDFL for identifying DFLs by transferring the IDR predictor RFPR-IDP to the DFL prediction. The RFPR-IDP was pre-trained with IDR data to learn the common features between IDRs and DFLs, and it was fine-tuned with the DFL data to capture the specific features of DFLs so as to be transferred into the DFL predictor TransDFL. Experimental results of two application scenarios (prediction of DFLs only in the IDRs and prediction of DFLs in the entire proteins) showed that TransDFL consistently outperforms the other exiting predictors with fewer false positives.