
 |
BioSeq-Analysis is a platform for DNA, RNA, and protein sequence analysis based on machine learning approaches |
For the convenience of the vast majority of experimental scientists, a step-by-step guide on how to use the BioSeq-Analysis web server to get their desired results without the need to follow the complicated mathematic equations is given below.
Visit the web-server by clicking the link at http://bliulab.net/BioSeq-Analysis/server and you will see the page as shown in Fig. 1. The three figures represent three sub web servers: DNA-Analysis, RNA-Analysis, and Protein-Analysis for DNA, RNA, and protein sequences, respectively.
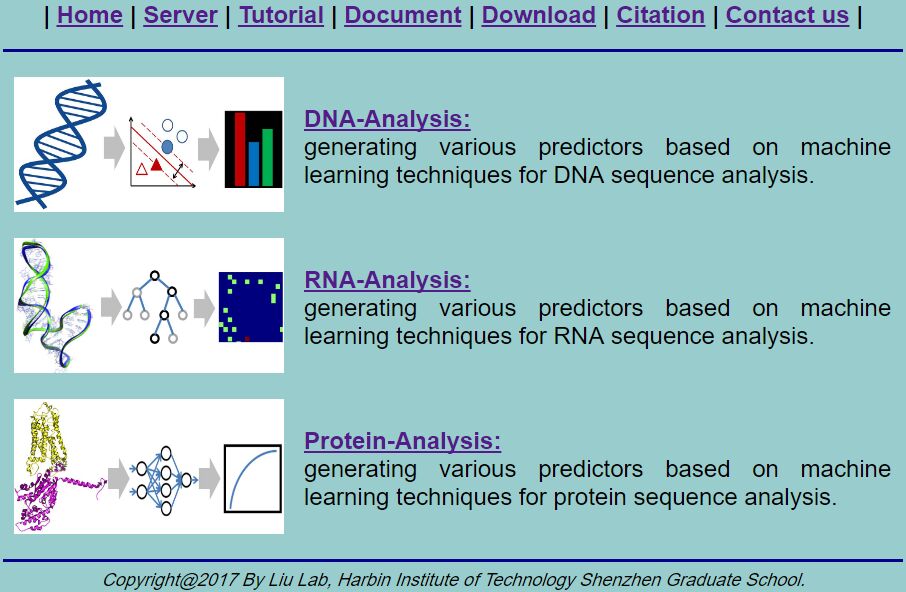
Fig. 1
Sequence analysis (Binary Classification)
Step 1. Take the DNA sequence for example. If you click the first picture (DNA Analysis) as shown in Fig. 1, then you will see the DNA-Analysis web server (Fig. 2).
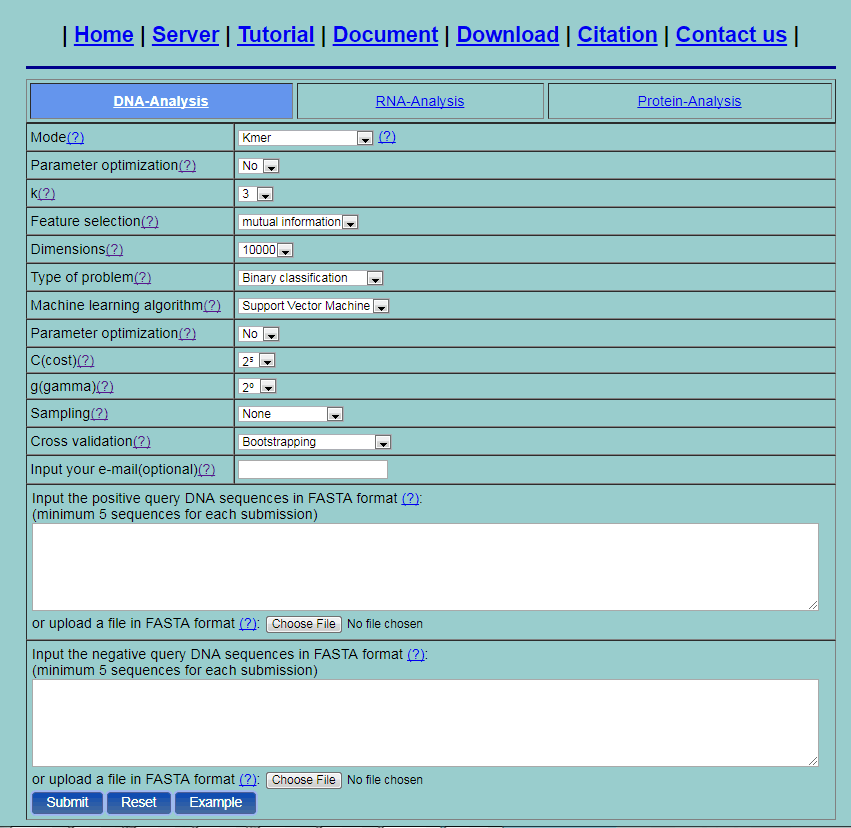
Fig. 2
Step 2. Select one method from the 20 methods listed in the pull-down menu, its corresponding parameters will be shown. You can click the “?” for the help information of various methods and their parameters. For example, if you select the Kmer method, you will see the page as shown in Fig. 2. The first "parameter optimazition" option is used for the feature extraction methods. Here we set the option as "YES" and then the parameters will be given as a range of values. If the feature vectors are too long, you can do feature selection by setting the feature selection and dimension options. Set the type of problem as "Binary classification". Then only two input boxes for positive dataset and negative dataset are shown. Two machine learning algorithms can be used: support vector machine and random forest. Select one algorithm, and its corresponding parameters will shown. The second "parameter optimazition" option is used for the machine learning algorithms. Here we set the option as "YES" and then the parameters will be given as a range of values. For binary classification problem, if any parameter optimization option is set, the "Performance measure" option is available. This option provides the performance measure used for selecting the optimal parameters. Here we set the option as "ACC". To deal with the problem of unbalance datasets, under sampling and oversampling are provided. Then set the cross validation method. Here three methods are provided: 5-fold cross validation, independent dataset test and bootstrapping. Here, select 5-fold cross validation. You can either type or copy and paste the query DNA sequences into the input boxes or directly upload your input data by clicking the Choose File button. The input sequences should be in the FASTA format. You can just click the button Example to input the built-in sequence examples with default parameter setting as shown by Fig. 3.
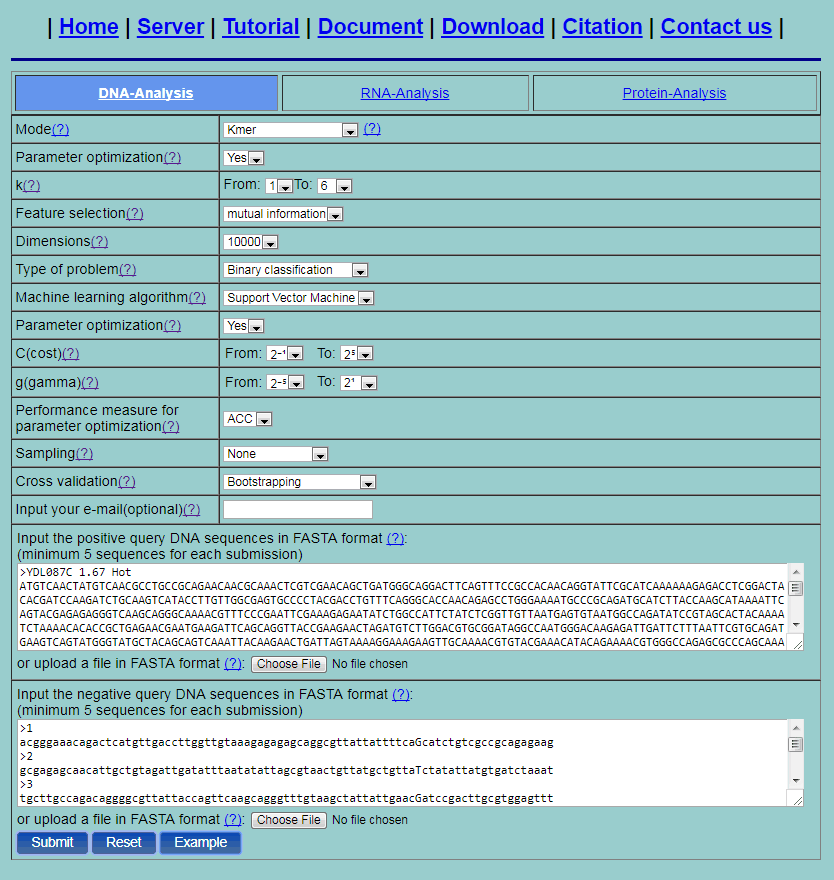
Fig. 3
Step 3. If you just click the button Example in the Step 2, and then click the Submit button, you will see a result page as shown in Fig.4. "Parameter Summary" shows the basic information of the mode and machine learning method used in the process and the corresponding parameters. After the "Parameter Summary" is the evaluation results. If 5-fold cross validation is used, it will show the performance measures of 5-fold cross validation. If the problem is binary classification problem, the ROC curve will be shown. Besides, you can click the Download button to download the feature vectors in a text file.

Fig. 4
Step 1. Take the DNA sequence for example. Enter the DNA-Analysis webserver shown in Fig. 1.
Step 2.Select one method from the 20 methods listed in the pull-down menu and its corresponding parameters will be shown. Take the "Kmer" method for example. Set the parameter optimazition as "No", and then the parameters of the mode will be a specific value. Set the type of problem as "Multiclass classification". Then the "Number of class" option will be shown. Set the "Number of class" as "3". Three input boxes will be provided for users to input datasets of three class respectively. Click the button Example to input the built-in sequence examples. Set the machine learning algorithm as support vector machine and its corresponding parameters. Set the parameter optimization as "No", and then the parameters of the algorithm will be a specific value. Select values for the parameters. Then set the cross validation method as 5-fold cross validation. The setting of parameters are shown as Fig. 5.
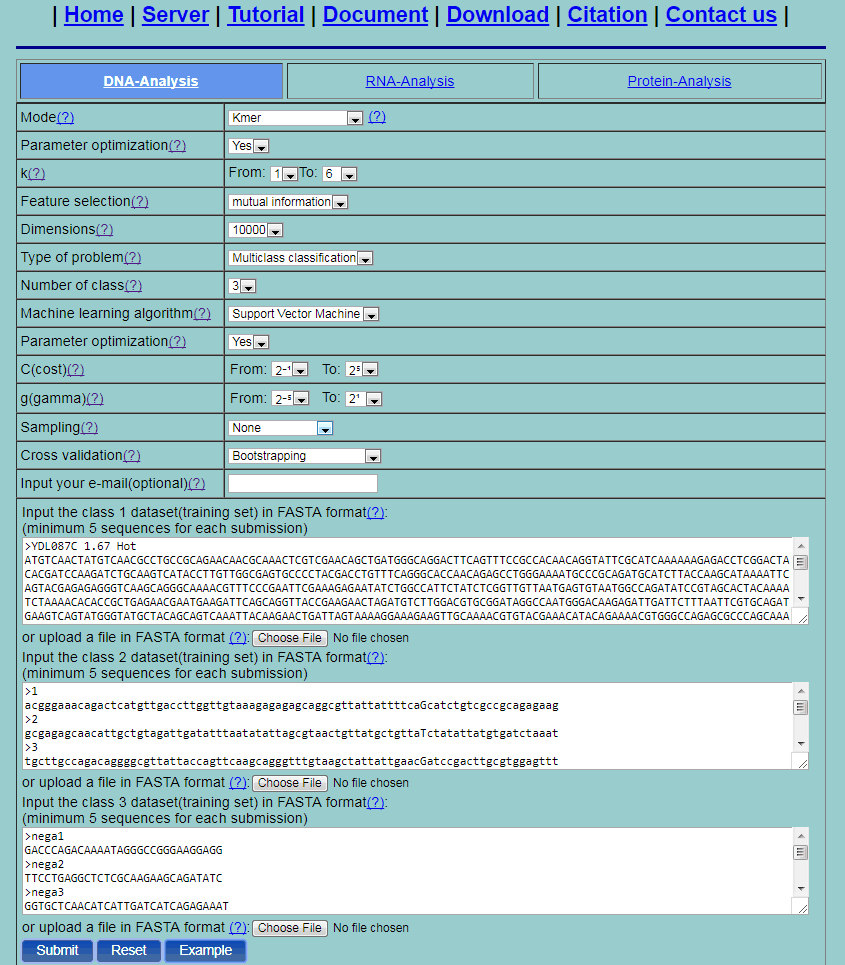
Fig. 5
Step 3. Click the Submit button, you will see a result page as shown in Fig.6. After the "Parameter Summary" is the evaluation results. For multiclass classification problem, only ACC is given. Besides, the confusion matrix is calculated. You can click the Download button to download the feature vectors in a text file.
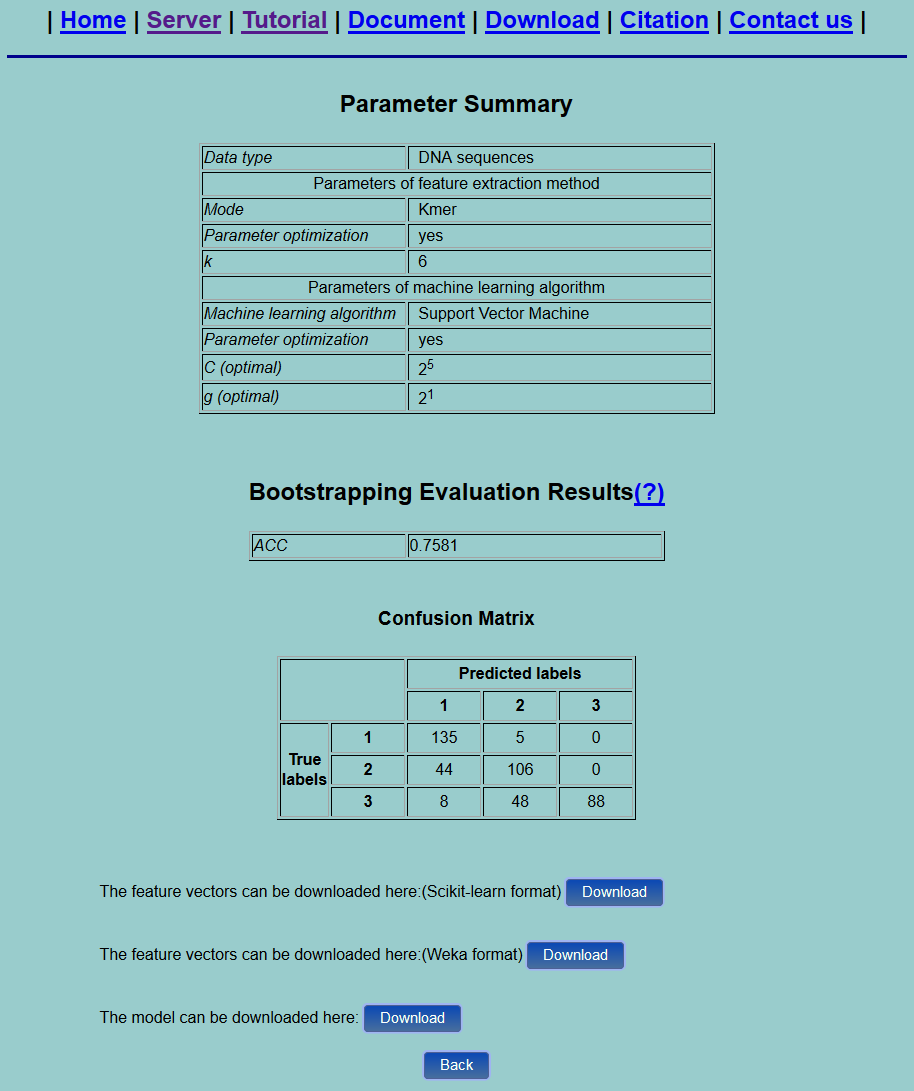
Fig. 6