 |
GraLTR-LDA:lncRNA-disease association prediction based on graph auto-encoder and Learning to Rank |
 |
GraLTR-LDA:lncRNA-disease association prediction based on graph auto-encoder and Learning to Rank |
Discovering the relationships between long non-coding RNAs (lncRNAs) and diseases is significant in the treatment, diagnosis and prevention of diseases. However, current identified lncRNA-disease associations are not enough because of the expensive and heavy workload of wet laboratory experiments. Therefore, it is greatly important to develop an efficient computational method for predicting potential lncRNA-disease associations. Previous methods showed that combining the prediction results of the lncRNA-disease associations predicted by different classification methods via Learning to Rank (LTR) algorithm can be effective for predicting potential lncRNA-disease associations. However, when the classification results are incorrect, the ranking results will inevitably be affected. We propose the GraLTR-LDA predictor based on biological knowledge graphs and ranking framework for predicting potential lncRNA-disease associations. Firstly, homogeneous graph and heterogeneous graph are constructed by integrating multi-source biological information. Then, GraLTR-LDA integrates graph auto-encoder and attention mechanism to extract embedded features from the constructed graphs. Finally, GraLTR-LDA incorporates the embedded features into the LTR via feature crossing statistical strategies to predict priority order of diseases associated with query lncRNAs. Experimental results demonstrate that GraLTR-LDA outperforms the other state-of-the-art predictors, and can effectively detect potential lncRNA-disease associations.
As shown in Fig.1, we treat the lncRNA-disease association prediction as a graph-based search task, similar as the searching task of searching associated movies for query actor in search engine. Graph-based knowledge storage is a kind of structured knowledge representation in knowledge graph. The current advanced search engines utilize the entity knowledge in the structured knowledge graph to find the entities associated with the query entities. For the lncRNA-disease association search task, the lncRNA-disease association graph is considered as biological knowledge graph.
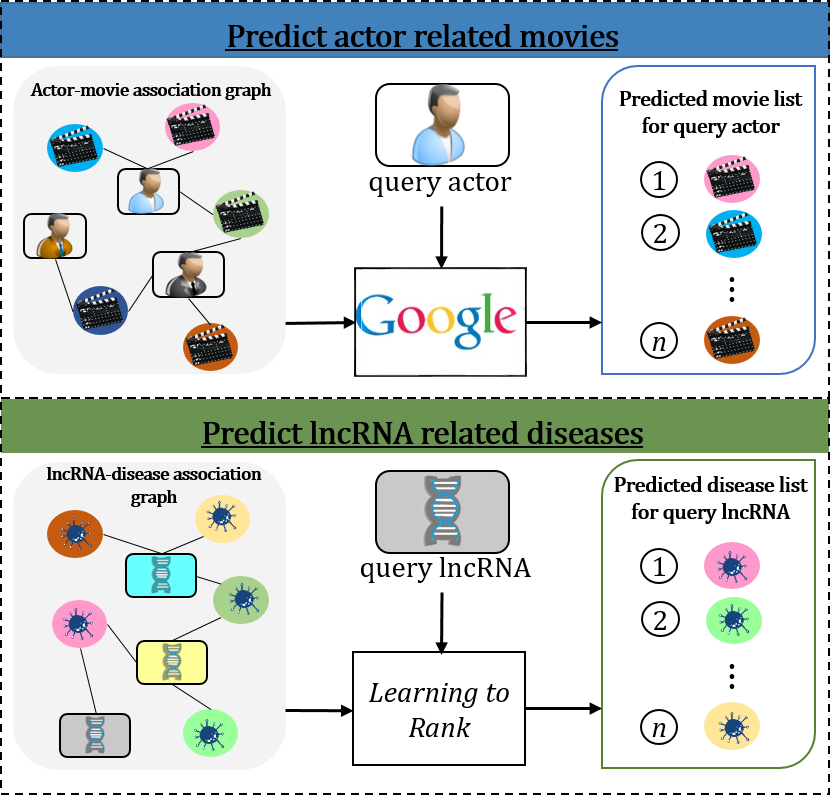
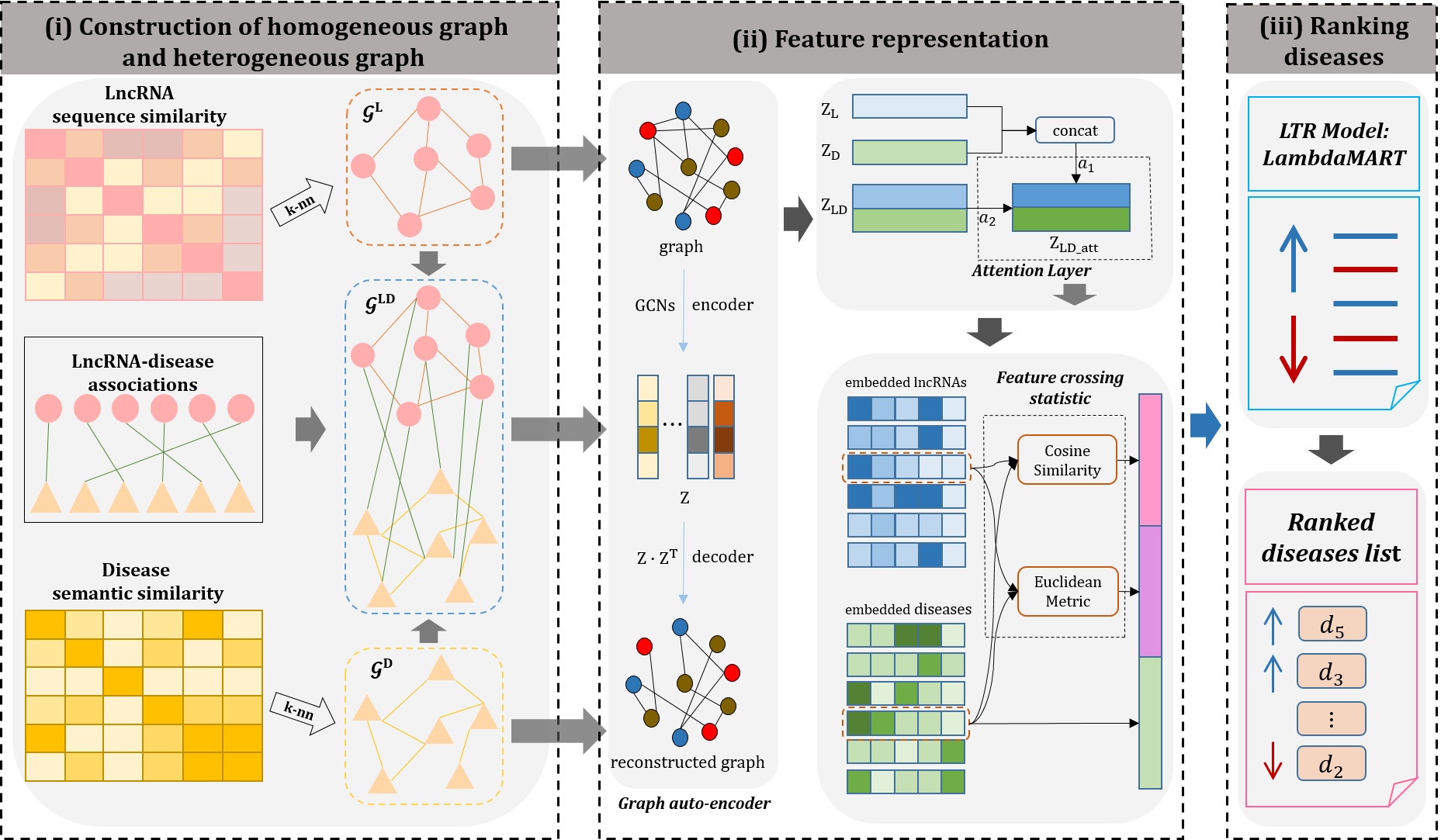
The flowchart of GraLTR-LDA model is shown in Fig.2, which contains three main steps, including construction of homogeneous graph and heterogeneous graph, feature representation, and ranking diseases.
| Datasets and source codes are available at: (GraLTR-LDA_main.rar) |
|
Citation: Liang, Q., Zhang, W., Wu, H. and Liu, B. LncRNA-disease association identification using graph auto-encoder and learning to rank. Briefings in Bioinformatics, 2023, 24(1): bbac539. |