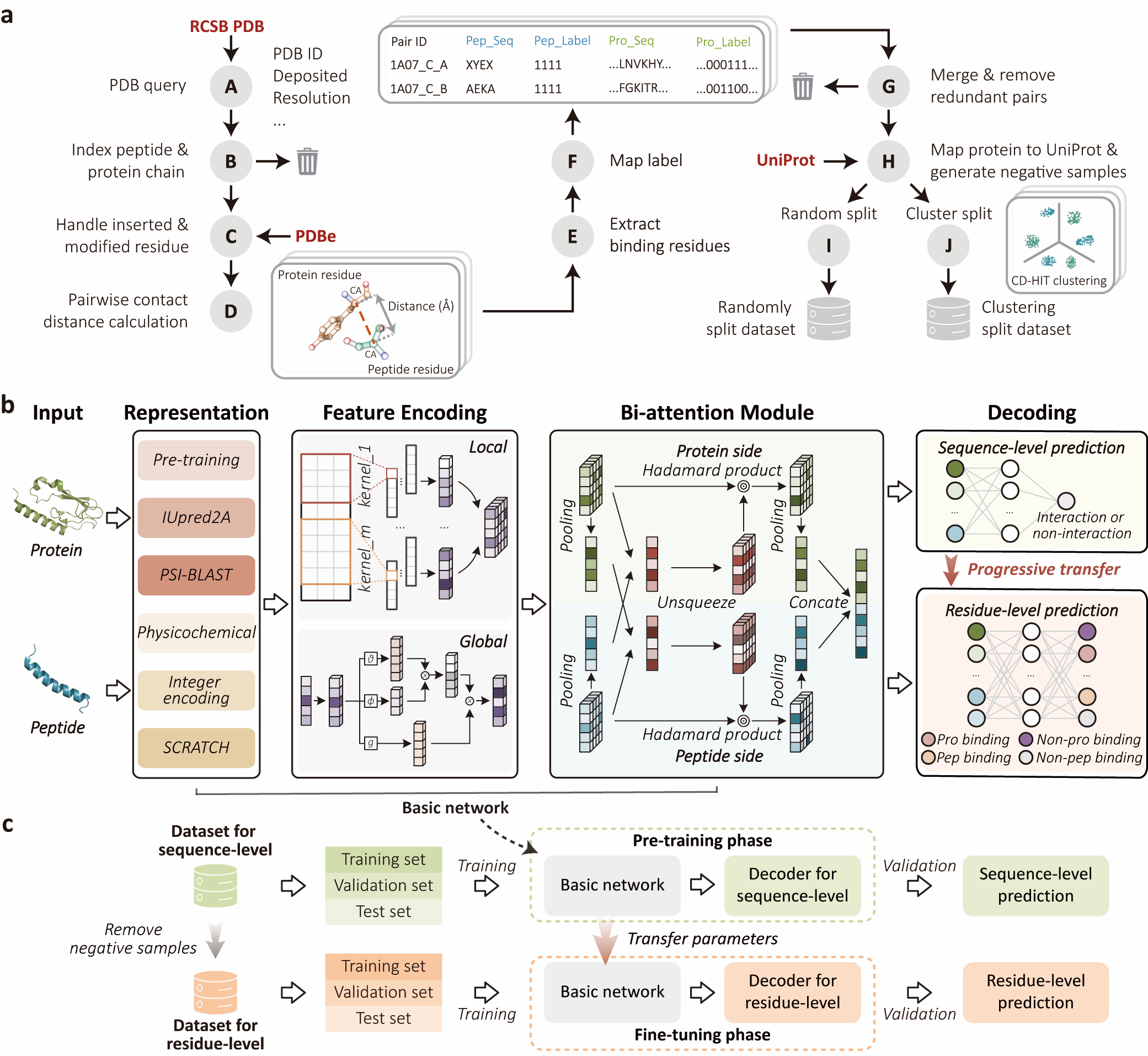
Introduction
Protein complex structural data is growing at an unprecedented pace, but its complexity and diversity pose significant challenges for protein function research. Although deep learning models have been widely used to capture the syntactic structure, word semantics, or semantic meanings of polypeptide and protein sequences, these models often overlook the complex contextual information of sequences. Here, we propose IIDL-PepPI, a deep learning model designed to tackle these challenges by employing data-driven and interpretable pragmatic analysis to profile peptide-protein interactions (PepPIs). IIDL-PepPI constructs bidirectional attention modules to represent the contextual information of peptides and proteins, enabling pragmatic analysis. It then adopts a progressive transfer learning framework to simultaneously predict PepPIs and identify binding residues in specific interactions, providing a solution for multi-level in-depth profiling. We validate the performance and robustness of IIDL-PepPI in accurately predicting peptide-protein binary interactions and identifying binding residues compared with state-of-the-art methods. We further demonstrate the capability of IIDL-PepPI in peptide virtual drug screening, binding affinity assessment, and complex residue flexibility prediction, which is expected to advance artificial intelligence-based peptide drug discovery and protein function elucidation.