 |
NCBRPred:predicting nucleic acid binding residues in proteins based on multi-label learning |
 |
NCBRPred:predicting nucleic acid binding residues in proteins based on multi-label learning |
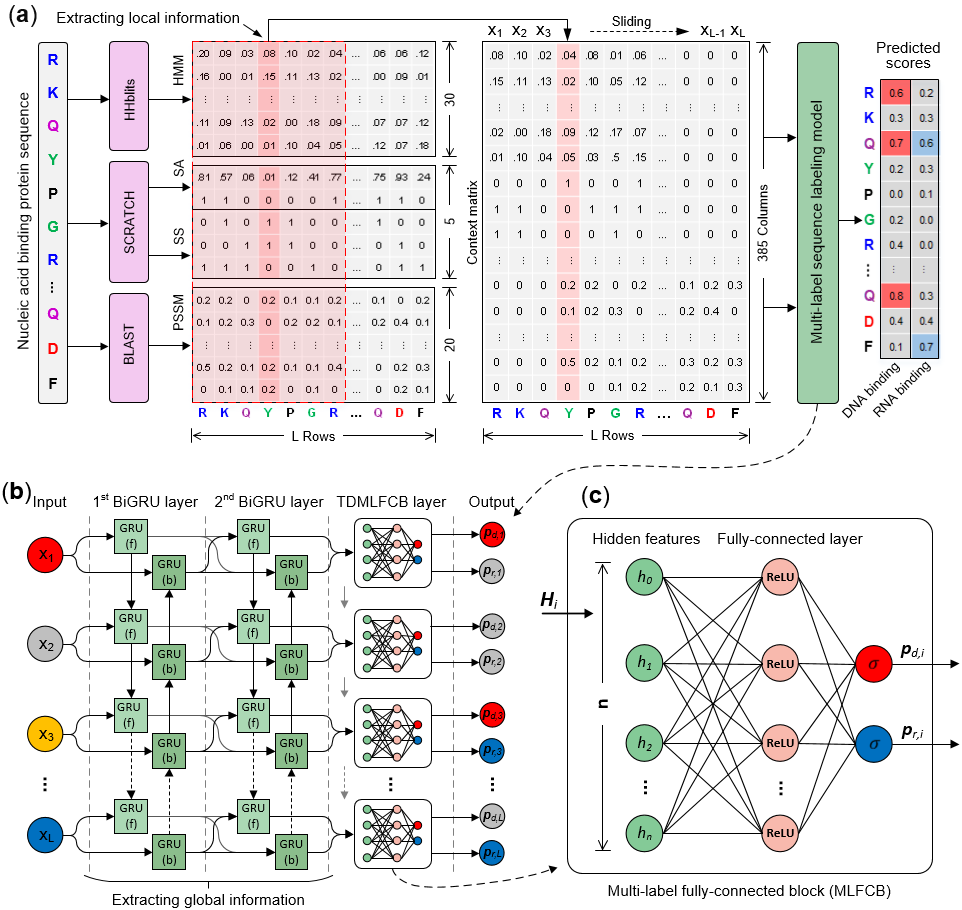
NCBRPred is a new sequence-based computational predictor for identifying DNA-binding residues and RNA-binding residues. In which multi-label learning framework and sequence labeling model were employed. It improved performance of nucleic acid binding residue prediction whlie maintaining a low cross-prediction rate, which is an important complement to existing methods. The framework and working process of NCBRPred are shown in Fig. 1. For more information please refer to our paper.
We acknowledge with thanks the following softwares used as a part of this server:
(1) PSIBLAST - Generation of the position specific scoring matrixes (PSSMs);
(2) NRDB90 - The non-redundant database for usage of PSIBLAST to generate PSSMs;
(3) HHBlits - Generation of the hidden Markov model (HMM) based evolutionary profiles;
(4) HHsuite database - The non-redundant database for uasage of HHblits to generate HMM profiles;
(5) SSpro and ACCpro - Prediction of protein secondary structure and solvent accessibility respectively;
(6) Biopython - Protein data preprocessing;
(7) Keras - Construction of the prediction model;
(8) Tensorflow - Backend of keras for computing.
The stand-alone packages of NCBRPred based on python 2.7 and python 3.7 can be download from below links:
[NCBRPred(Python2) | NCBRPred(Python3) | README.txt]
Note: For the example of the command line of the stand-alone package and the guide of configuring the stand-alone package please refer to the above README file.
Upon the usage of this server the users are requested to use the following citation:
Jun Zhang, Qingcai Chen, Bin Liu*. NCBRPred: predicting nucleic acid binding residues in proteins based on multi-label learning. Briefings in Bioinformatics,2021,22(5):bbaa397.