About PL-search
Protein remote homology detection is a fundamental and important task in the analysis of protein structure and function. Many search methods have been proposed to improve the detection of re-mote homologues and the accuracy of ranking lists. The Position Specific Scoring Matrix (PSSM) profile and Hidden Markov Model (HMM) profile can contribute to improving the performance of state-of-the-art search methods.
In this paper, we trace profile-link information used to construct the PSSM or HMM profiles in order to propose a Profile-Link-based search method (denoted PL-search). In PL-search, more robust profile links are constructed through the double-link and iterative extending strategies, and an accu-rate similarity score of sequence pairs is calculated from the two-level Jaccard distance for remote homologues. We tested our method on the classic and updated versions of the SCOP benchmark datasets. Our results show that whether HHblits, JackHMMER or PSI-BLAST are used, PL-search significantly improves the search performance in terms of ranking quality as well as the number of detected remote homologues.
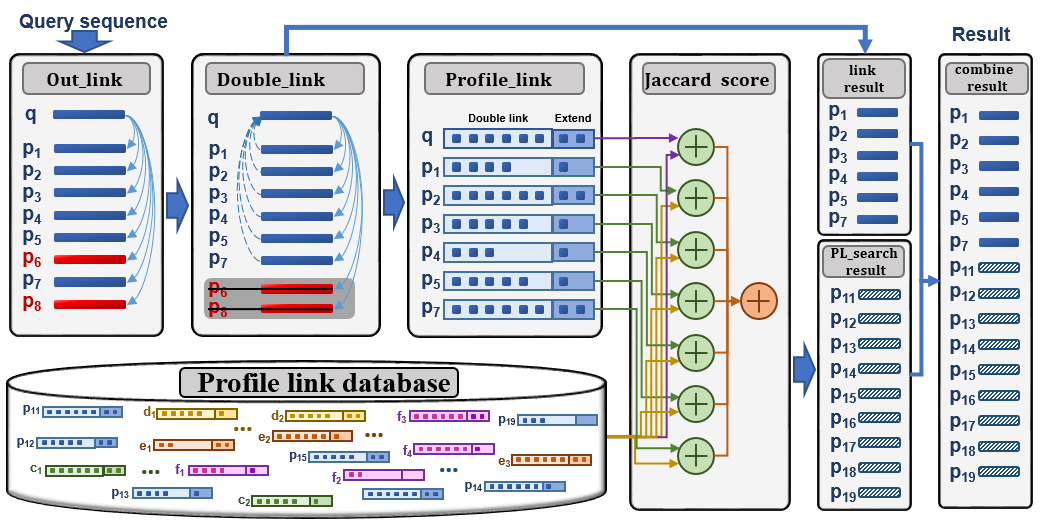
Tested on the classic version and updated version of SCOP benchmark datasets, experimental results show that whatever HHblits, JackHMMER or PSI-BLAST it base on, PL-search significantly improves the search performances not only in ranking quality but also in the number of detected remote homology protein sequences.
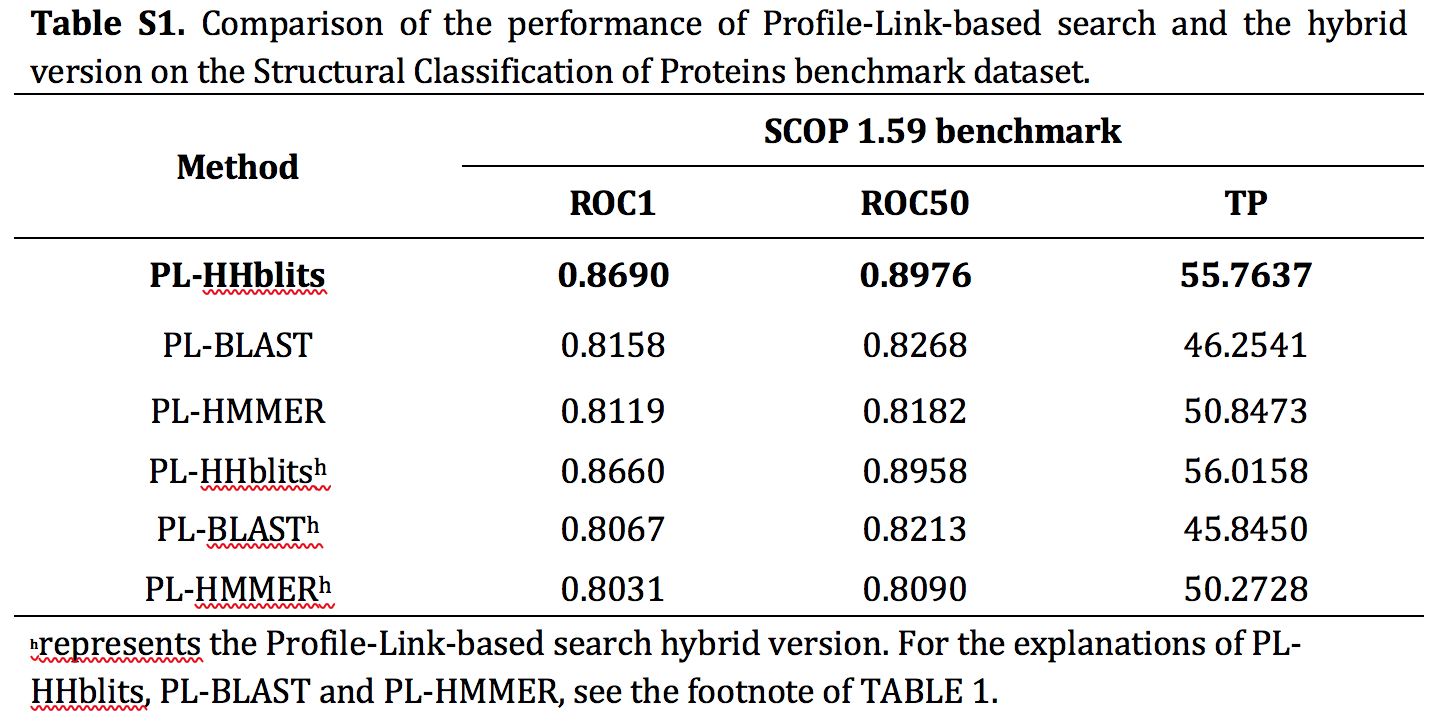
For the web server, constructed profile-link databases lead the in-link for new protein sequences cannot be obtained. Therefore, we propose a hybrid version of PL-search for the web server, which exhibits a little accuracy loss (Table S1). In the web server, to calculate the similarity of protein pairs, the first level of the Jaccard distance is calculated by out-link and profile link, and the calculated manner of the second level of the Jaccard distance is retained. The final ranking list is constructed from search results and out-link instead of the double-link (cf. Eq. S1 and Eq. S2).
The similarity score of sequence pairs from the two-level Jaccard distance is calculated in the hybrid version using Equation S1:
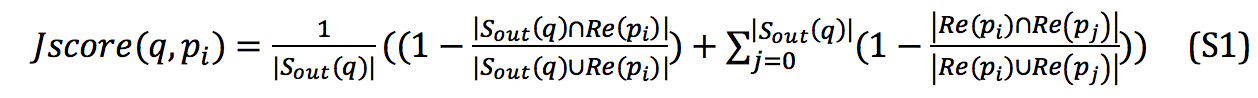
The final ranking list in the hybrid version is calculated with Equation S2: