Feature analysis
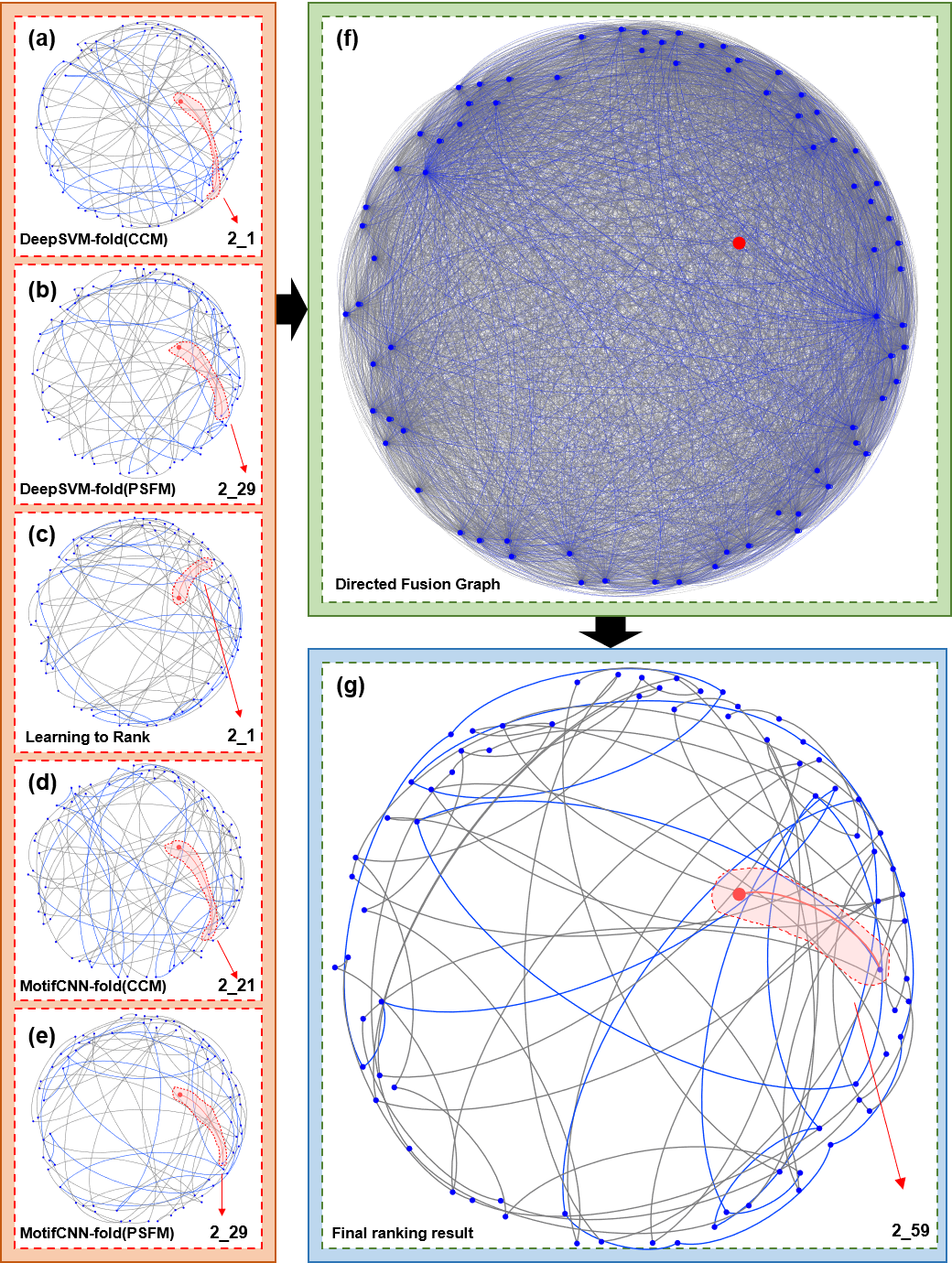
In order to further explore the reasons why the ProtFold-DFG is able to accurately detect the protein folds, a query protein 1bdo-d1bdo from protein fold 2_59 (SCOP ID) is selected as an example, and the prediction results of ProtFold-DFG are visualized in Fig. 5, from which we can see the following: i) According to the top hits in the five basic ranking lists shown in Fig. 5 (a)-(e), the fold type of the query protein is incorrectly predicted by the five computational predictors. ii) DFG is able to fuse the five ranking lists by considering the local and global relationships among proteins (see Fig. 5 (f)). iii) Performed on the DFG, the PageRank algorithm is able to re-rank the proteins in the DFG and correctly predict the fold of the query protein 1bdo-d1bdo as 2_59 (see Fig. 5 (g)). These results are not surprising, although the top hits in the basic ranking lists of the five predictors are incorrect, these errors can be corrected by fusing these complementary ranking lists and analyzing their relationships comprehensively. Briefly, ProtFold-DFG treats the proteins the scientific literature and considers the most closely linked by others as the matched result for the query.
Reference
1. Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. In: Third international AAAI conference on weblogs and social media. 2009.