 |
iCircDA-LTR:identification of circRNA-disease associations based on Learning to Rank |
 |
iCircDA-LTR:identification of circRNA-disease associations based on Learning to Rank |
Circular RNAs (circRNAs) are a class of non-coding RNAs forming closed loop structures without 5' and 3' polyadenytaled tails. More and more studies demonstrate that circRNAs are serving as major regulators in various cellular processes and associated with multiple diseases. Therefore, identifying circRNA-disease associations can help understand functions of newly discovered circRNAs and explore new disease therapy strategy.
Motivated by the similarities between circRNA-disease association identification task and topic-document pair search task (Fig.1), we proposed a new predictor called iCircDA-LTR. It has the following advantages: (i) The ranking framework based on Learing to Rank (LTR) is able to fuse different methods in a supervised manner, overcoming the disadvantages of both classification methods and recommendation methods while reserving their advantages; (ii) The ranking framework is able to globally consider the relationships among all the candidate diseases, and reduce the false positives, especially for the top ones. Therefore, iCircDA-LTR is practically useful for predicting the diseases associated with new circRNAs; (iii) iCircDA-LTR is a ranking framework. Other predictors can be incorporated to further improve its performance; (iv) For the convenience of researches, a web server of iCircDA-LTR was established to help explore new associations between diseases and query circRNAs.

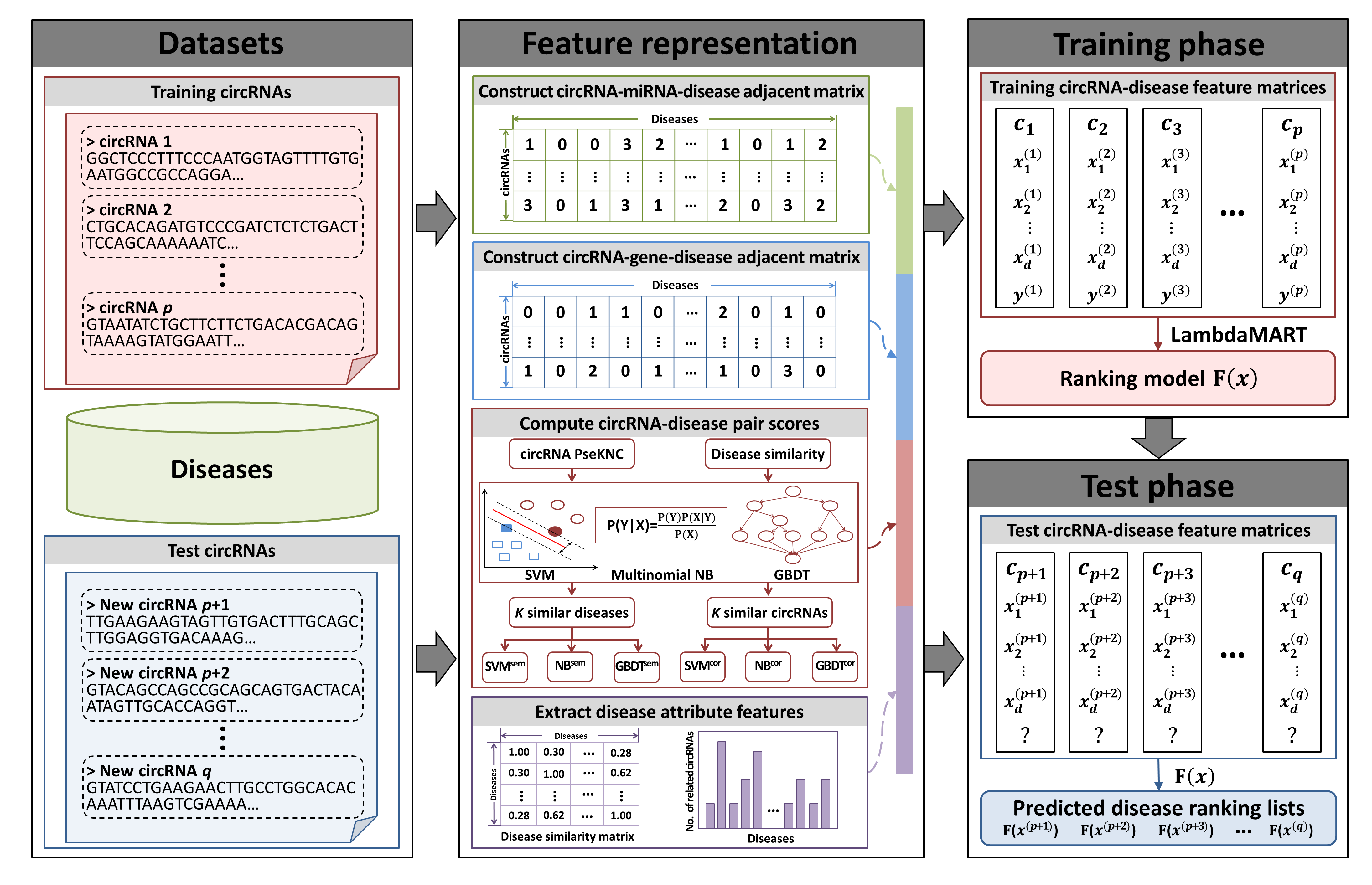
There are three main steps in iCircDA-LTR (Fig.2): (i) Feature representation. Each training or test circRNA is embedded into a feature matrix based on heterogeneous information, where the rows represent the pair feature vectors of target circRNA with different candidate diseases. The feature vector of each circRNA-disease pair can be described from four parts including: circRNA-miRNA-disease pair score, circRNA-gene-disease pair score, different machine-learning-method-based pair scores and disease attribute features; (ii) LTR training phase. The feature matrices for training circRNAs are given as input to train the LTR model. A listwise algorithm of LTR, LambdaMART is applied to learn a ranking model; (iii) LTR test phase. For a query circRNA, a ranking list of candidate diseases can be predicted by the learned ranking model, where the disease associated with the query circRNA will be ranked in the top.
CONTACT US
Bin Liu
bliu@bliulab.net
School of Computer Science and Technology, Beijing Institute of Technology, China.
Copyright@ By Liu Lab, Beijing Institute of Technology.
网站备案号: 粤ICP备19041859号-1