|
iLncDA-LTR:identification of lncRNA-disease associations by learning to rank |
 |
iLncDA-LTR:identification of lncRNA-disease associations by learning to rank |
Identifying the associations between lncRNA and disease is helpful for the treatment and diagnosis of complex diseases. The existing computational methods mainly focus on the identification of associations between known lncRNA and known disease. However, with the application of high-throughput sequencing in lncRNA research, more and more lncRNAs have been detected. Predicting diseases related with newly-found lncRNAs has not yet been fully explored in existing methods. Therefore, there is an urgent need for developing a powerful computational method to predict diseases related with newly-found lncRNAs.
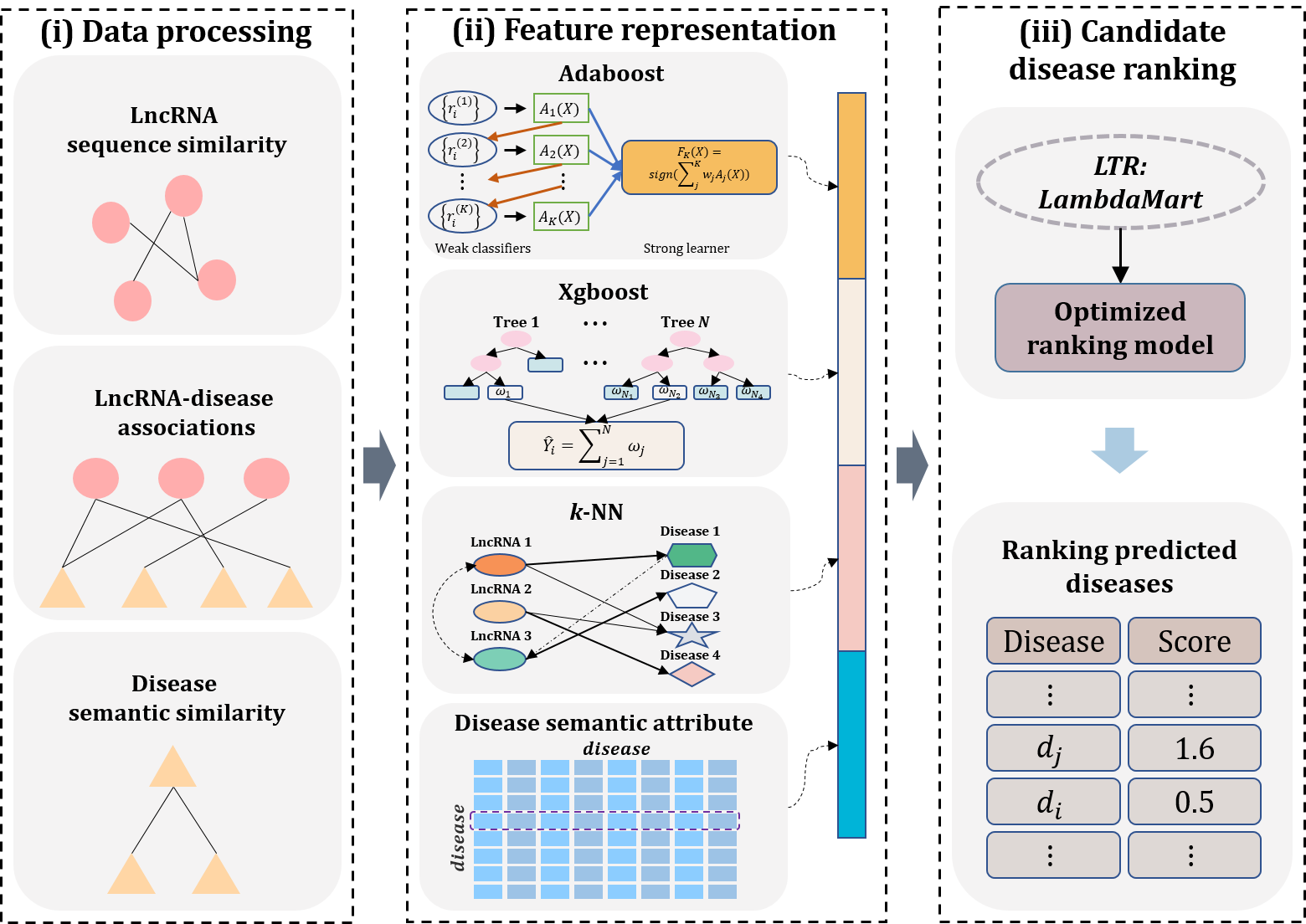
In this paper, we propose a Learning to Rank (LTR)-based method called iLncDA-LTR to predict diseases related with newly-found lncRNAs. iLncDA-LTR treats the problem of identifying associations between newly-found lncRNAs and diseases as information retrieval task, in which newly-found lncRNAs and diseases are regarded as queries and documents, respectively. For a given newly-found lncRNA (query), iLncDA-LTR integrates multiple relevant information into LTR for predicting candidate diseases associated with query lncRNA. The flowchart of iLncDA-LTR model is shown in Fig.1, which contains three main steps, including data processing, feature representation and candidate disease ranking.