|
iSnoDi-LSGT:identifying snoRNA-disease associations based on local similarity constraint and global topological constraint |
 |
iSnoDi-LSGT:identifying snoRNA-disease associations based on local similarity constraint and global topological constraint |
Small nucleolar RNA (snoRNA) is a type of small non-coding RNAs with 60-300 nucleotides in length. In the process of revealing the functions of snoRNAs, more and more evidences indicated that snoRNAs play crucial roles in the emergence and spread of diseases.
Various snoRNA databases were constructed to reveal the functions of snoRNAs, such as snoRNAdb, snOPY, SNORic, etc. Although these databases revealed the functions of snoRNA in details, the research on snoRNA-disease associations was still in its infancy. The main reason is that current studies of snoRNA-disease associations are mainly based on biological experiments. In recent years, MNDR (Mammalian ncRNA-Disease Repository) v3.0 database integrated considerable experimentally supported and predicted ncRNA-disease associations, and provided an opportunity to efficient identify disease-related snoRNAs by computational methods.
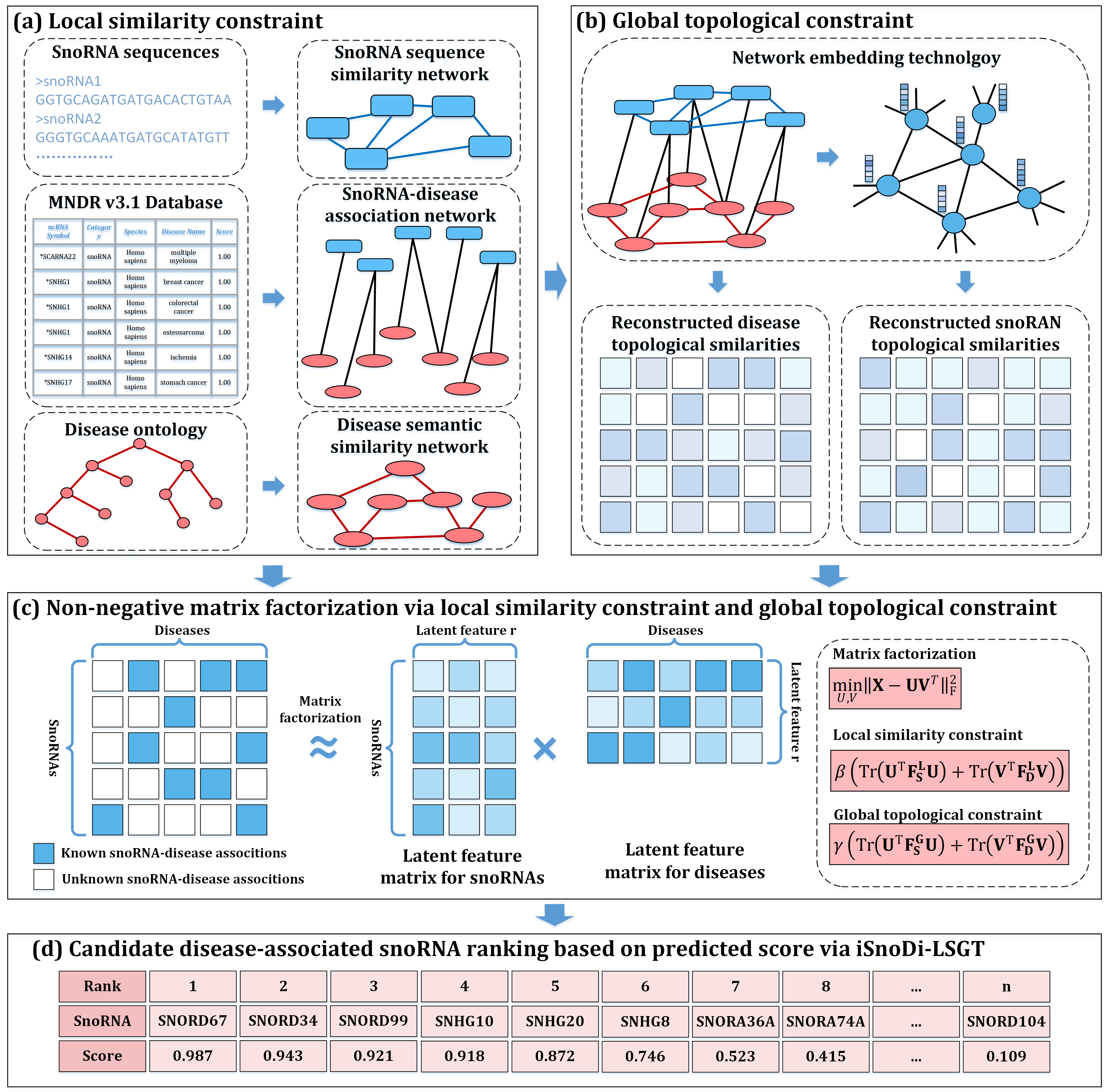
In this study, a computational method named iSnoDi-LSGT(as Figure.1) is proposed to identify unknown snoRNA-disease associations via local similarity constraint and global topological constraint. The iSnoDi-LSGT predictor has the following advantages: (i) To the best knowledge of ours, iSnoDi-LSGT predictor is the first computational predictor for snoRNA-disease association identification; (ii) The local similarity constraint, global topological constraint, snoRNA and disease global topological features are incorporated into iSnoDi-LSGT predictor to accurately identify the snoRNA-disease associations.