|
idenMD-NRF:a ranking framework for miRNA-disease association identification |
 |
idenMD-NRF:a ranking framework for miRNA-disease association identification |
MicroRNAs (micro ribonucleic acid, miRNAs) are endogenous non-coding RNAs (ncRNA) containing about 23 nucleotides, which exist in the cells and play a vital role in regulating kinds of life activities. Accumulating evidences have indicated that miRNAs are closely associated with the emergence and progression of complicated diseases. Therefore, identifying miRNA-disease associations is a significant task for revealing pathogenic mechanism of complicated diseases.
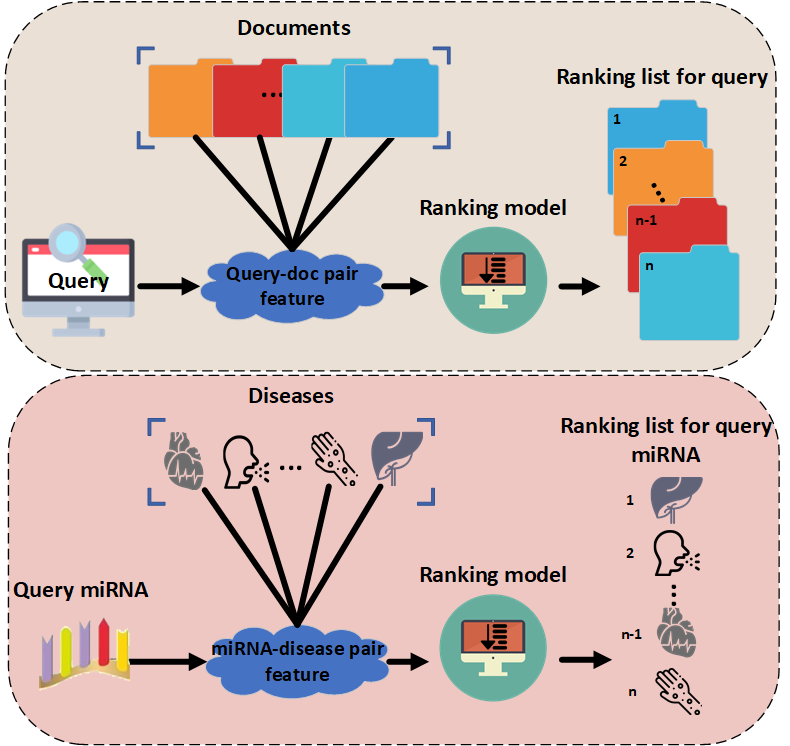
Information retrieval task is similar with the task of identifying miRNA-related diseases (as Fig.1). For the process of document retrieval task, given a novel query, the ranking model can rank associated documents and then return the top-k documents. The same procedure can be implemented for predicting diseases associated with query miRNAs through a trained ranking model, where the miRNAs and diseases are analogous to queries and documents respectively.
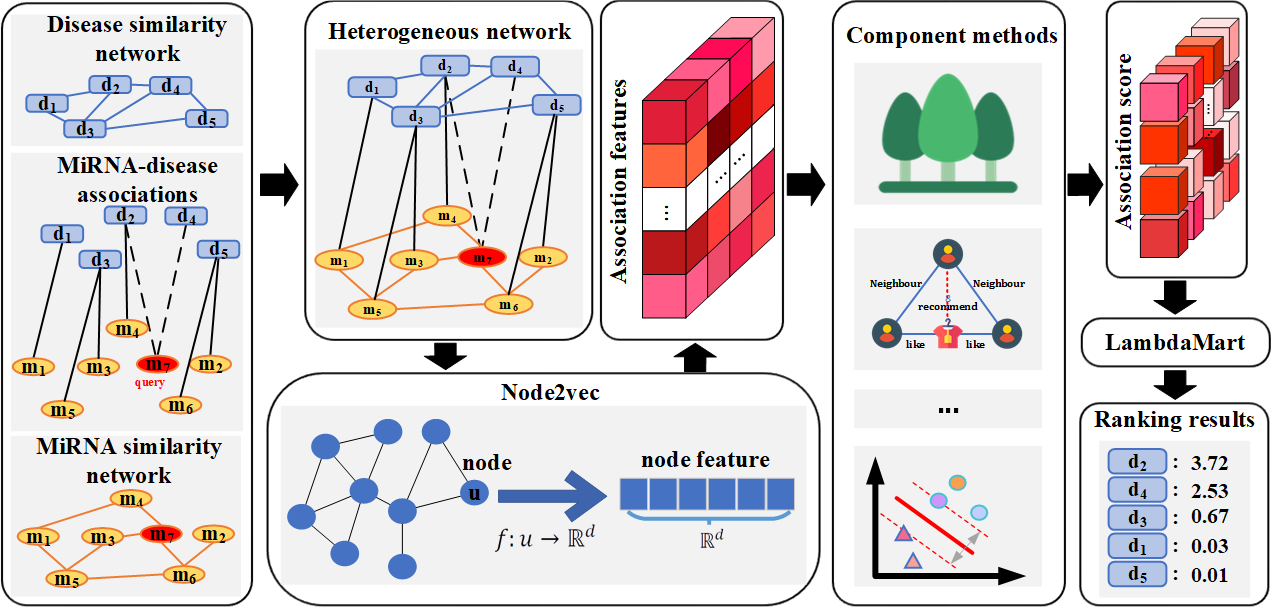
Motivated by the successful application of Learning to Rank (LTR) to information retrieval, we propose a predictor called idenMD-NRF(as Fig.2) to identify new miRNA-disease associations by a ranking framework. Compared with existing methods, it has following contributions: (i) idenMD-NRF can work for both two application scenarios: detecting missing associations between known miRNAs and diseases, and predicting diseases associated with newly detected miRNAs. (ii) idenMD-NRF is an inclusive ensemble ranking framework. Therefore, complementary predictors can be integrated for further performance improvement. (iii) It is biased that most existing methods treated unknown associations as negative samples. However, some potential positive associations may be in the unknown sample set. idenMD-NRF can consider the top-ranked samples, and greatly weaken the negative impact of unknown samples. (iv) A new deep learning technique with strong graph representation ability, node2vec, is employed by idenMD-NRF to capture the high-level association features