 |
DeepDRBP-2L:a new genome annotation predictor for identifying DNA-binding proteins and RNA-binding proteins using Convolutional Neural Network and Long Short-Term Memory |
 |
DeepDRBP-2L:a new genome annotation predictor for identifying DNA-binding proteins and RNA-binding proteins using Convolutional Neural Network and Long Short-Term Memory |
DNA-binding proteins (DBPs) and RNA-binding proteins (RBPs) are two kinds of crucial proteins, which are involved in various cellule activities and associated with many diseases. Furthermore, some proteins (DRBPs) can bind to both DNA and RNA, which play important roles in gene expression. Accurate identification of DBPs, RBPs and DRBPs is critical for both theoretical research (understanding the protein functions and the transcription process) and real world application (drug design). In this regard, various computational predictors have been proposed. However, they were constructed only to detect one of DBPs or RBPs, resulting a large number of DBPs, RBPs and DRBPs to be misclassified and exposing to the risk of overestimation.
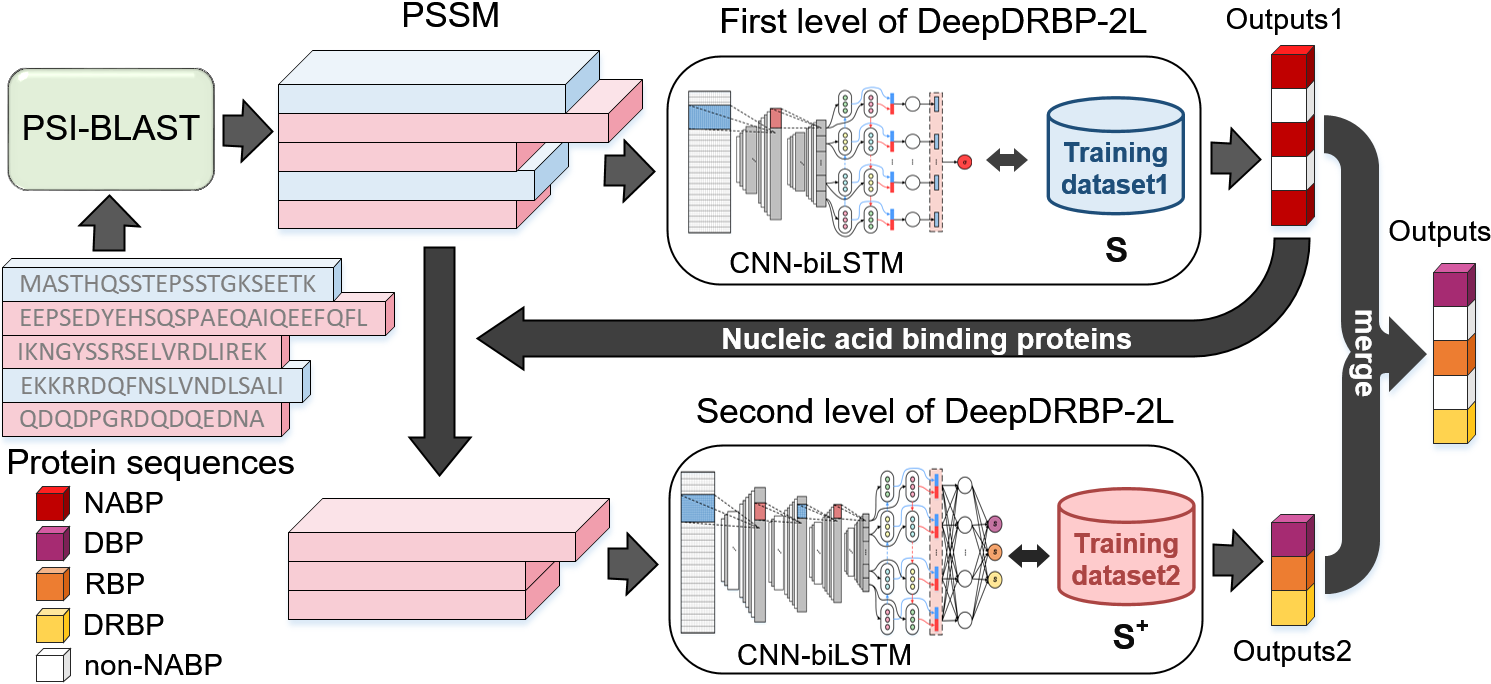
In this study, we proposed a new computational predictor called DeepDRBP-2L for identifying DBPs, RBPs and DRBPs. Compared with other existing methods, it has the following advantages: 1) DeepDRBP-2L is the first computational predictor that is able to identify DBPs, RBPs and DRBPs; 2) DeepDRBP-2L employs a two-level framework, the first level detects the nucleic acid binding proteins (DNA/RNA-binding proteins, NABPs), and the second level further identifies DBPs, RBPs and DRBPs by scoring, which successfully overcomes the over-estimation problem; 3) The deep learning techniques were employed to improve the recognition accuracy, which are able to capture the different characteristics between NABPs and non-NABPs or DBPs and RBPs; 4) DeepDRBP-2L is able to accurately detect the DBPs and RBPs at the proteome scale, which will be a useful tool for protein function annotation. The framework and working process of DeepDRBP-2L are shown in Fig.1.
Keras-2.0.8
Theano-0.9.0
Numpy-1.11.2
Biopython-1.68
Scikit-learn-0.19.2
The benchmark dataset used in this study can be download from below link:
[Benchmark dataset, Sequences]
The independent dataset constructed in this study can be download from below link:
[Independent dataset, Sequences]
CONTACT US
Bin Liu
bliu@bliulab.net
School of Computer Science and Technology, Beijing Institute of Technology, China.
网站备案号: 粤ICP备19041859号-1
Copyright@ By Liu Lab, Beijing Institute of Technology.