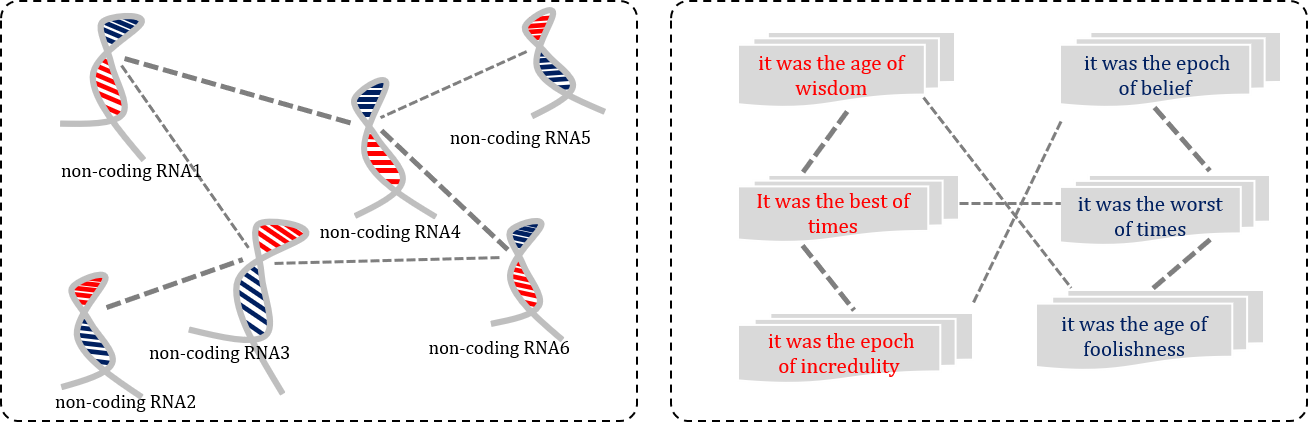
Biological data type
Homogeneous biological sequence similarities
For the homogeneous biological sequence similarities, the queries and the retrieved samples are homogeneous.
(left) Non-coding RNA similarity analysis, which is a homogeneous biological sequence analysis task. (right) Non-coding RNA and disease association identification, which is a heterogeneous biological sequence analysis task.
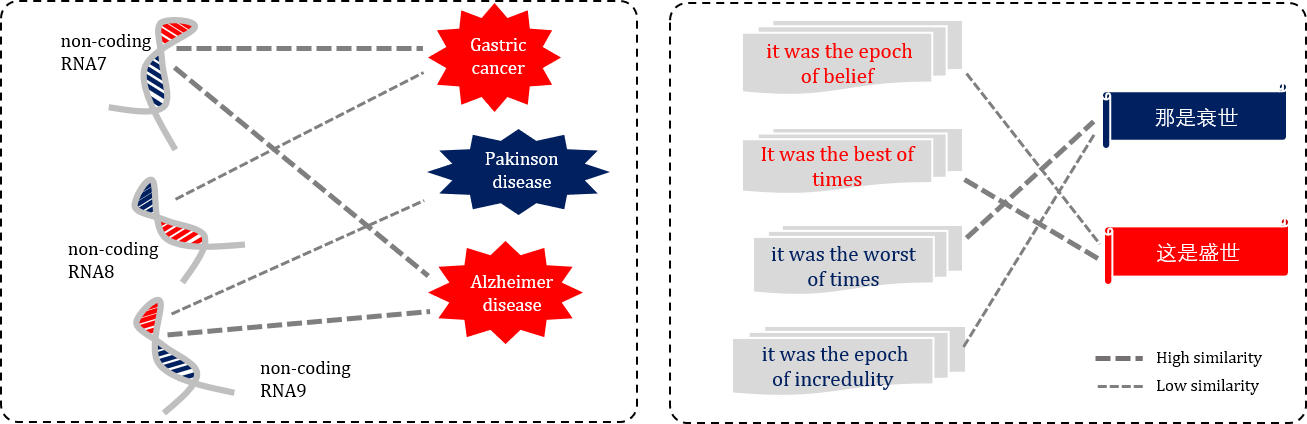
Heterogeneous biological sequence similarities
For the heterogeneous biological sequence similarities, the queries and the retrieved samples are heterogeneous.
(left) Text matching task, which is a homogeneous language analysis task. (right) Machine translation task, which is a heterogeneous language analysis task.
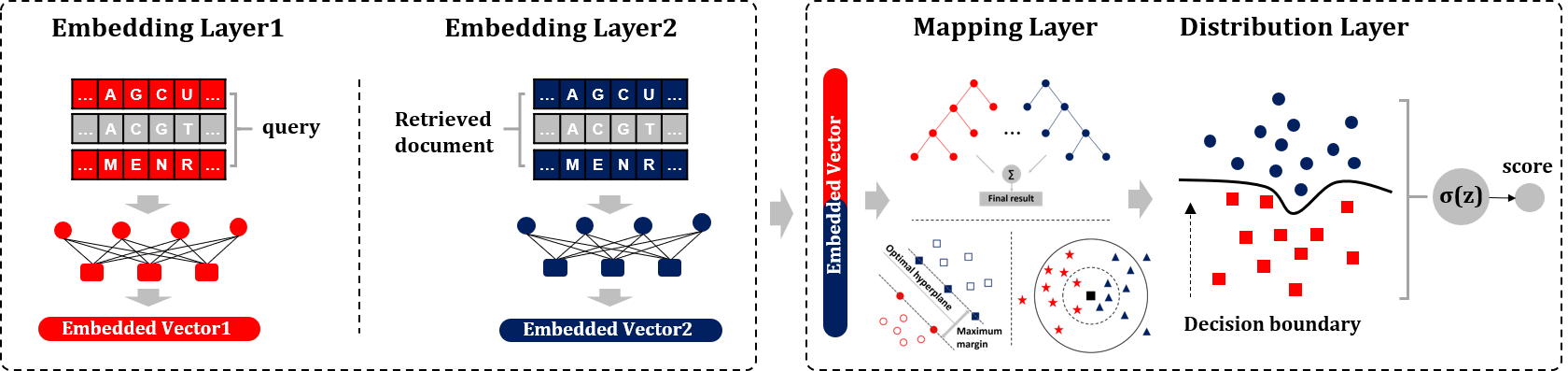
Biological sequence similarities calculation methods
Distribution methods
Distribution methods fully consider the spatial correlation of input pairs, and show good generalization ability for modelling different types of data. [1]
Representation methods
The representation methods employ the Siamese architecture to encode the sentences, which can be applied to analyse the biological sequence similarities. [1]

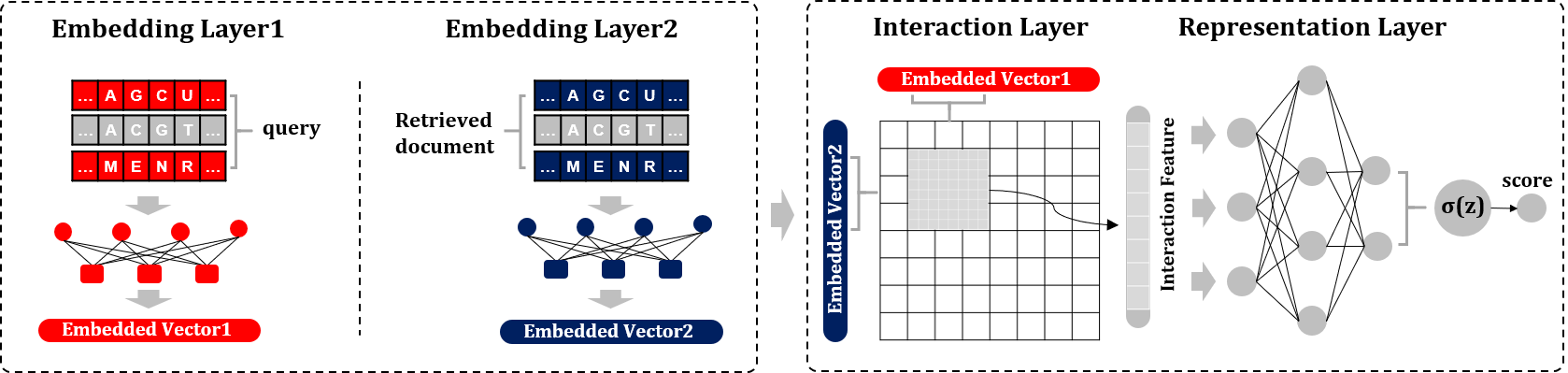
Interaction methods
Interaction methods employ the hierarchical deep architecture to learn the semantics from the local interaction matrix of query and retrieved documents, which is suitable for comprehensively learning the associations between biological sequences. [1]
Reference:
[1] Chandrasekaran D, Mago V. Evolution of Semantic Similarity—A Survey. ACM Comput Surv. 2021;54(2):41. doi: 10.1145/3440755.
The input data should be in the BLS format. Detailed information of the BLS format is introduced in the followings:
Required format of input biological sequences.
Please enter the biological sequences in FASTA format.
Example:
>5www_A
GHHHHHHMQAALLRRKSVNTTECVPVPSSEHVAEIVGRQLGMVLWIYKWFKPDGRLTDEQIADGMVGMLFPPFYIKTPVRGEEPIFVVTGRKEDVAMAKREILSAAEHFSMIRAS
Required format of the input vectors.
Please enter the feature vectors in following format (similar to FASTA format).
>vec_name1
vec_val1 vec_val2 vec_val3 ... vec_valn
Example:
>Data_A_ID:0
0.045 0.027 0.035 0.030 0.039 0.023 0.006 0.032 0.030 0.021 0.045 0.024 0.023 0.029 0.035 0.035 0.199 0.168 0.158
Required format of the input labels.
Please enter the associations in list format.
Example:
0 0
1 1
2 0
2 1
3 3
3 31
4 3
Interaction with BioSeq-BLM
Format conversion
If users want to generate feature vectors for input biological sequences, the pipeline software, BioSeq-BLM is recommended.
Based on biological language models, BioSeq-BLM can extract features representing linguistics attributes and biological attributes of biological sequences.
Users can serve the feature vectors generated by BioSeq-BLM as the input of BioSeq-Diabolo after a simple format conversion.