BioSeq-Diabolo:
biological sequence similarity analysis using Diabolo.
biological sequence similarity analysis using Diabolo.

The stand-alone package, manual and datasets of BioSeq-Diabolo are listed as follows.
The stand-alone package of BioSeq-Diabolo for Windows and Linux.
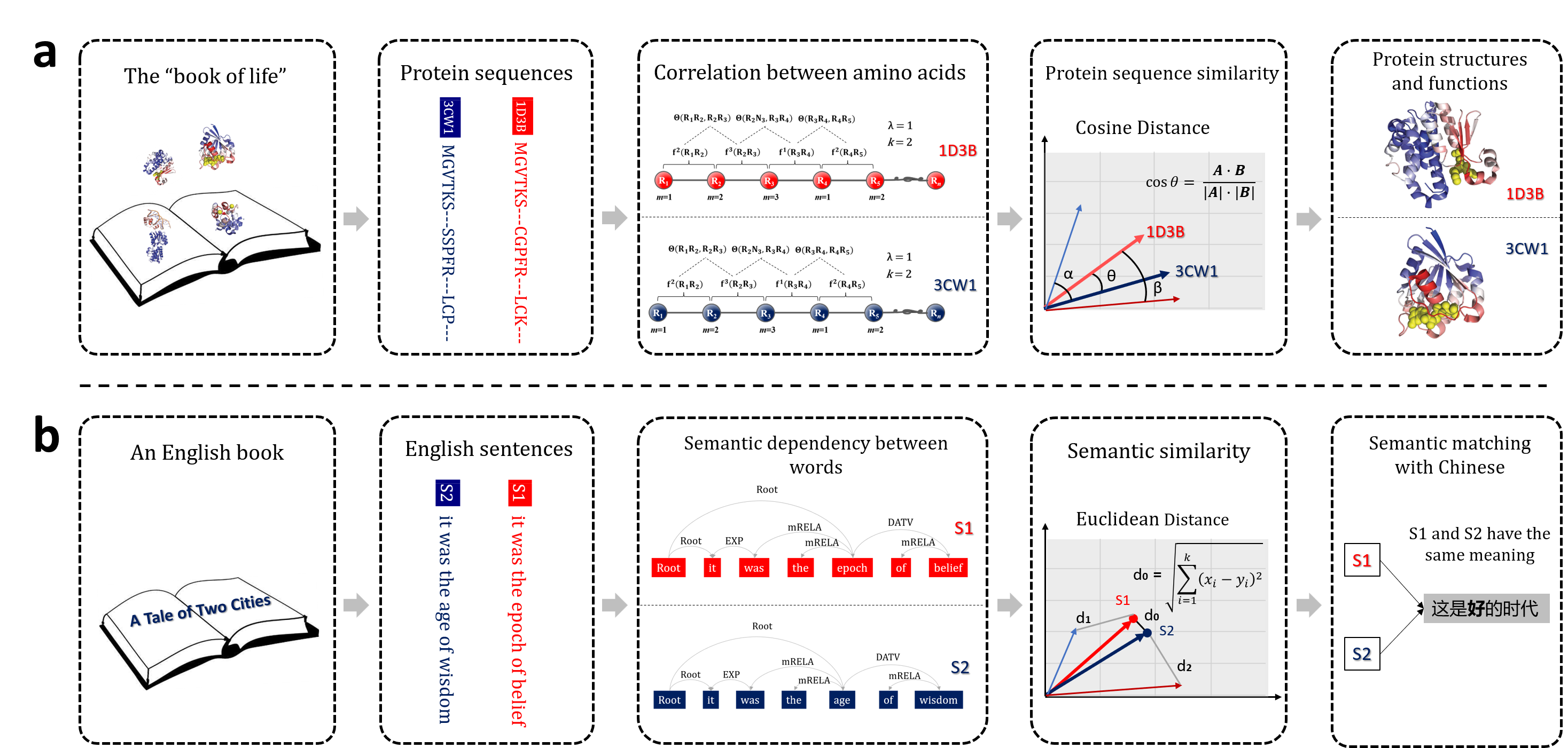
The related softwares are listed as follows. They are intelligent software tools for different tasks. BioSeq-BLM [1], BioSeq-Analysis2.0 [2] and BioSeq-Analysis [3] are designed for biological sequence feature extraction and predictors construction. And BioSeq-Diabolo can automatically construct the computational predictors for biological sequence similarity analysis, evaluate the performance, and analyze the results. BioSeq-Diabolo is an important updated version of BioSeq-BLM focusing on the homogeneous and heterogeneous biological sequence similarity analysing, which is beyond the reach of any exiting software tool or platform. The biological sequence features extracted by BioSeq-BLM can be served as the input of BioSeq-Diabolo, while the biological sequence similarity scores calculated by BioSeq-Diabolo can also be regard as the input features of BioSeq-BLM’s classification and sequence labelling models. The users are able to use these software tools for their own tasks and aims.
[1] Li H-L, Pang Y-H, Liu B. BioSeq-BLM: a platform for analyzing DNA, RNA and protein sequences based on biological language models. Nucleic Acids Research. 2021;49(22):e129-e. doi: 10.1093/nar/gkab829.
[2] Liu B, Gao X, Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Research. 2019;47(20):e127-e. doi: 10.1093/nar/gkz740.
[3] Liu B. BioSeq-Analysis: a platform for DNA, RNA, and protein sequence analysis based on machine learning approaches. Briefings in Bioinformatics 2019;20(4):1280-1294.
Hong-Liang Li, et al. BioSeq-Diabolo: biological sequence similarity analysis using Diabolo. PLOS Computational Biology, 2023, 19(6): e1011214.