 |
ProtDet-CCHProtein remote homology detection by combining Long Short-Term Memory and ranking methods |
 |
ProtDet-CCHProtein remote homology detection by combining Long Short-Term Memory and ranking methods |
As one of the most challenging tasks in sequence analysis, protein remote homology detection has attracted a great deal of interest. Methods based on discriminative models and ranking approaches have achieved the-state-of-the-art performance, and these two kinds of methods are complementary. However, the integration framework of combining the discriminative and ranking methods has never been explored.
In this study, three LSTM models have been used to construct the predictors for protein remote homology detection, including ULSTM, BLSTM, and CNN-BLSTM. They are able to automatically extract the local and global sequence order information. Combined with PSSMs, the CNN-BLSTM achieved the best performance among the three LSTM-based models. We named this method as CNN-BLSTM-PSSM. Finally, a new method called ProtDet-CCH was proposed by combining CNN-BLSTM-PSSM and a ranking method HHblits. Tested on a widely used SCOP benchmark dataset, ProtDet-CCH achieved an ROC score of 0.998, and an ROC50 score of 0.982, significantly outperformed other existing state-of-the-art methods. Experimental results on two updated SCOPe independent datasets showed that ProtDet-CCH can achieve stable performance. Furthermore, our method can provide useful insights for studying the features and motifs of protein families and superfamilies. It is anticipated that ProtDet-CCH will become a very useful tool for protein remote homology detection.
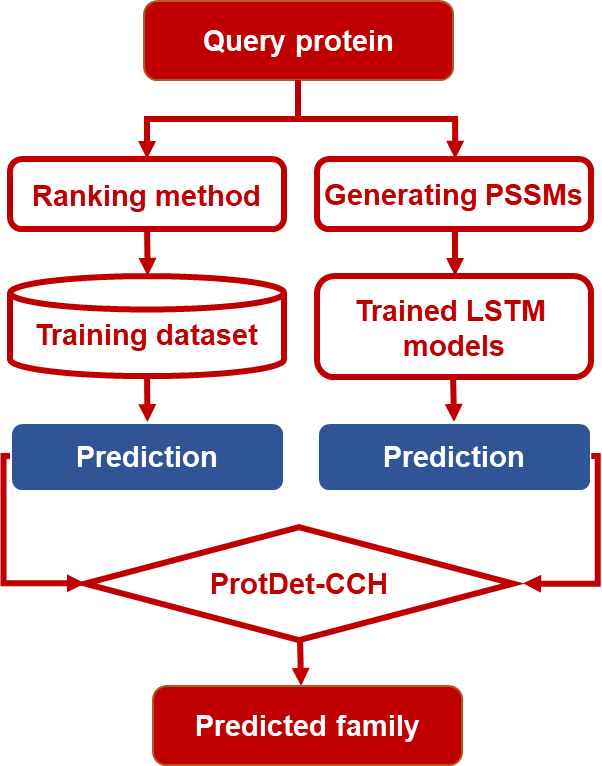
The supplementary files consists of the main python programs and the used datasets. To use these programs or datasets, please download following files:
[ProtDet-CCH] [README]
[SCOP-benchmark dataset,Sequences] [SCOPe-2747,Sequences] [SCOPe-102,Sequences]
ProtDet-CCH depends on the following toolkits:
Keras-2.0.6
Theano-0.9.0
Numpy-1.11.2
Biopython-1.68
Please cite the following papers when using the programs or datasets at this website:
Liu B, Li S. ProtDet-CCH: Protein remote homology detection by combining Long Short-Term Memory and ranking methods, IEEE/ACM Transactions on Computational Biology and Bioinformatics 2019;16:1203-1210.