|
ProtDet-CCHProtein remote homology detection by combining Long Short-Term Memory and ranking methods |
 |
ProtDet-CCHProtein remote homology detection by combining Long Short-Term Memory and ranking methods |
ProtDet-CCH web server is constructed by combining CNN-BLSTM-PSSM and a ranking method HHblits, which is compatible with most major browsers, and the parallel speed-up is implemented. It offers an open and interactive web service, which accepts query sequences in FASTA format and returns the search results in a user-friendly manner. For the convenience of the experimental scientists, a step-by-step guide on how to use the ProtDet-CCH web server is given below.
Visit the web server by clicking the link at http://bliulab.net/ProtDet-CCH/server and you will see the page as shown in Fig. 1. The Microsoft Edge and Google Chrome browsers are recommended.
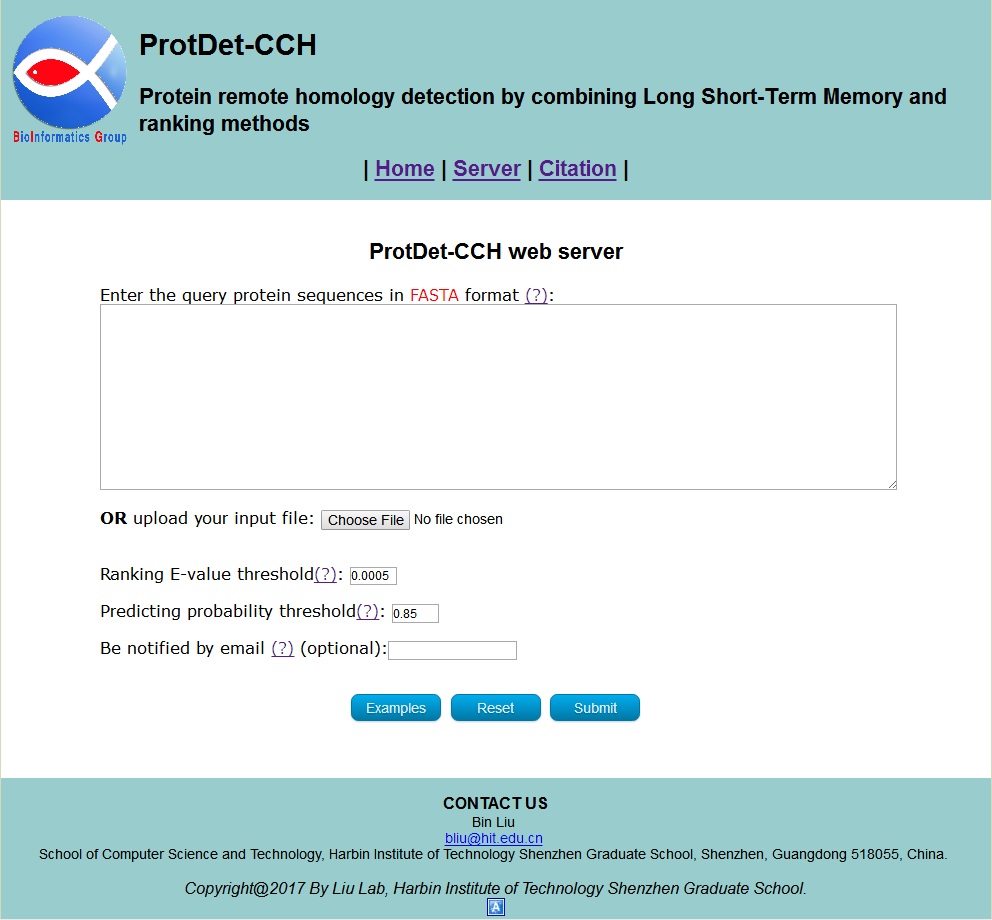
STEP 1: Input the query protein sequences
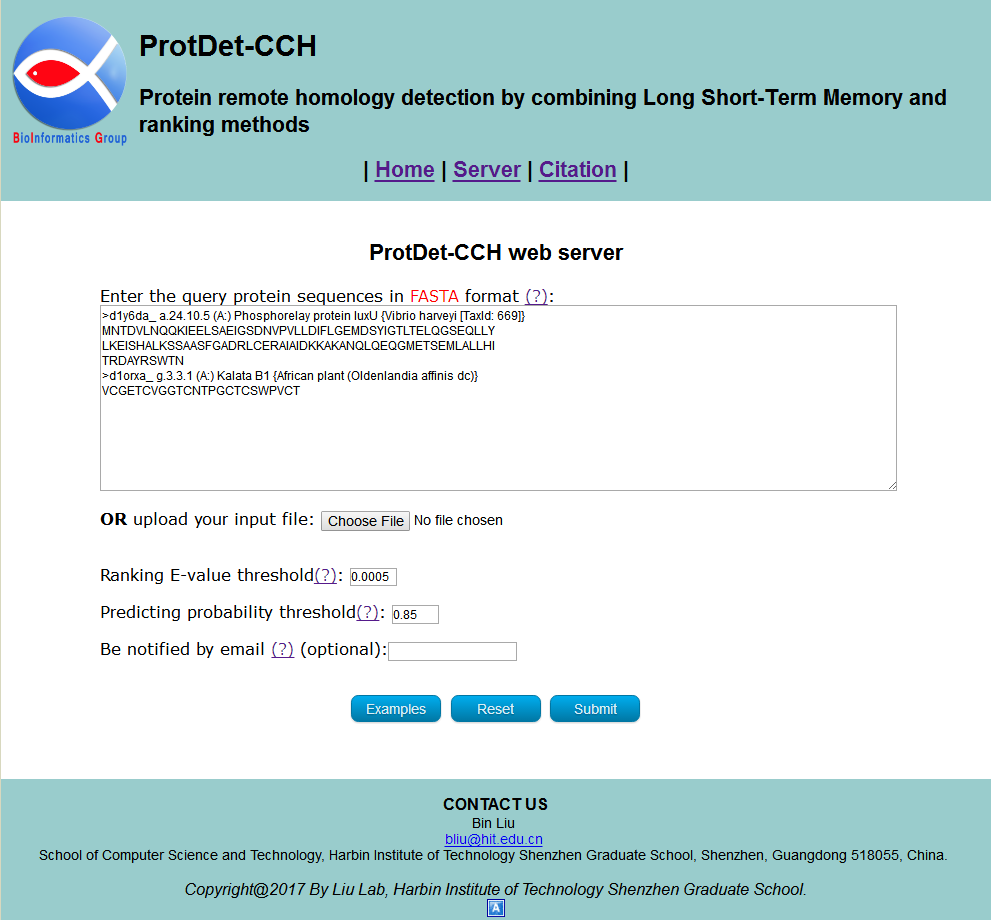
You can directly enter/paste the query protein sequences into the input box, or upload them via file by clicking the Choose File button. All the input sequences should be in the FASTA format. A sequence in FASTA format consists of a single line beginning with the symbol ">" and multiple lines of amino acids data. You can click the Examples button to automatically input the built-in sequence examples and the default parameter as shown in Fig 2, and click the Reset button to empty all input sequences and parameters.
STEP2: Set the parameters
The ranking E-value threshold and predicting probability threshold are necessary, which are thresholds for results output. If the minimum E-values in search result of HHblits smaller than ranking E-value threshold ProtDet-CCH will select the superfamily of the protein with the minimum E-values in search result of HHblits as final output result. If not, check the predicted results of 102 CNN-BLSTM-PSSM classifiers. If there are predicted probabilities greater than predicting probability threshold ProtDet-CCH will select the predicted result with maximum probability as final output result. If not, ProtDet-CCH will select the superfamily of the protein with the minimum E-values in search result of HHblits as final output result. The default value of ranking E-value threshold is 0.0005 and the default value of predicting probability threshold is 0.85, which are optimized for most users. Besides, the Email address is optional, please input your valid Email address so that we can send the results to you.
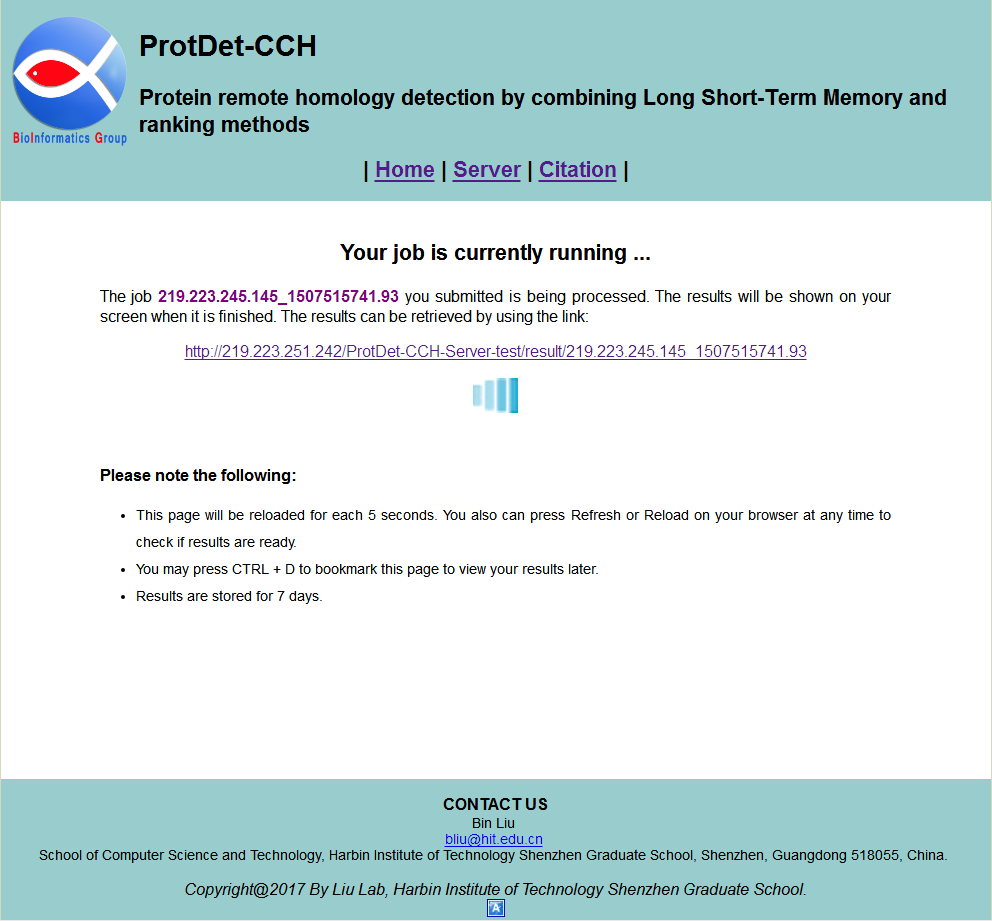
STEP 3: Submit your queries
If you enter query protein sequences, and then click the Submit button, you will see a processing page as shown in Fig. 3. Your job is being processed. The results will be shown on your screen when it is finished. You can also close this browser window, and reload the results by using the link.
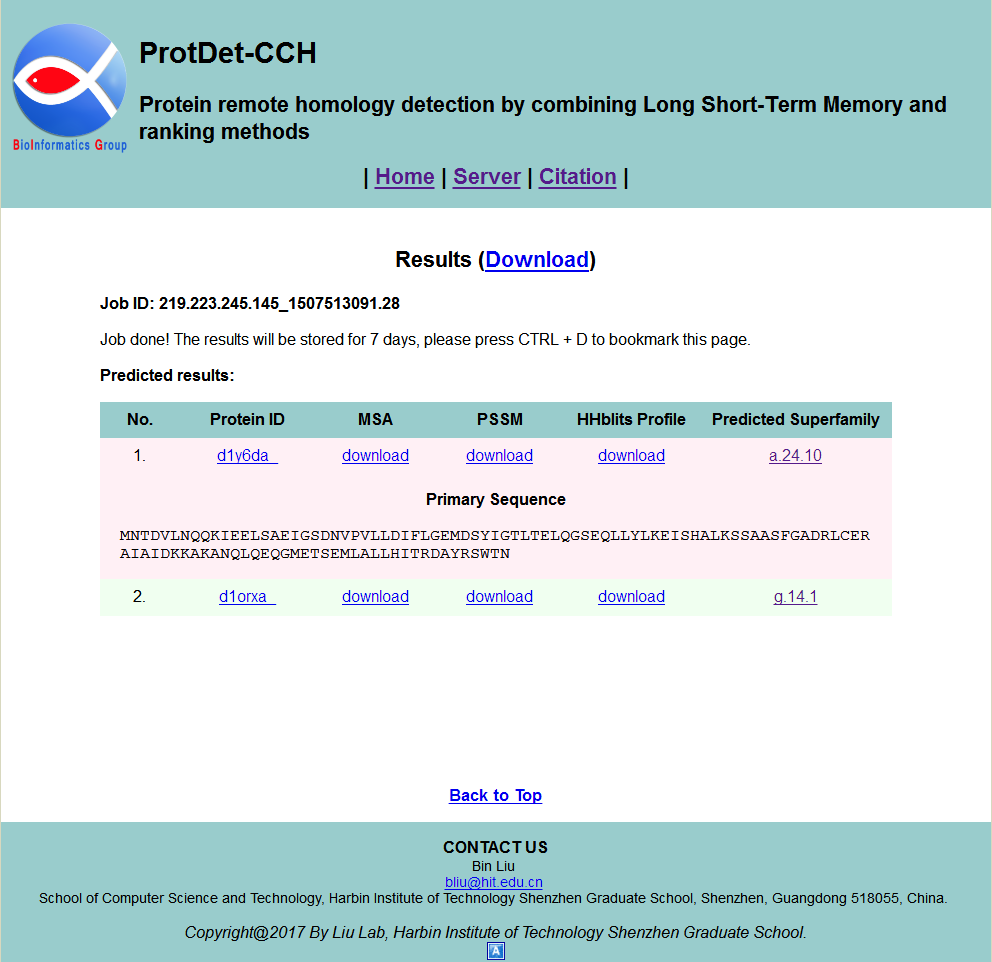
STEP 4: View results
The results will be shown on your screen when it is finished, as shown in Fig. 4. The results will be stored for 7 days, and you can click the Download link to download it. You can find the protein primary sequence by clicking the Protein ID button in Visualization column of the table. Corresponding Multiple Sequence Alignment (MSA), Position Specific Scoring Matrix (PSSM) and HHblits Profile can be download by clicking the corresponding download buttons in Visualization column of the table. The more detailed information about target superfamily of query protein can be found by clicking the Predicted Superfamily button at bottom of this cell.